Exploring the Impact of Batch Size on Deep Learning Artificial Intelligence Models for Malaria Detection
- PMID: 38868293
- PMCID: PMC11167577
- DOI: 10.7759/cureus.60224
Exploring the Impact of Batch Size on Deep Learning Artificial Intelligence Models for Malaria Detection
Abstract
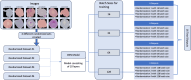
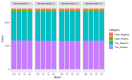
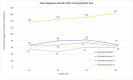
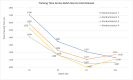
Introduction Malaria is a major public health concern, especially in developing countries. Malaria often presents with recurrent fever, malaise, and other nonspecific symptoms mistaken for influenza. Light microscopy of peripheral blood smears is considered the gold standard diagnostic test for malaria. Delays in malaria diagnosis can increase morbidity and mortality. Microscopy can be time-consuming and limited by skilled labor, infrastructure, and interobserver variability. Artificial intelligence (AI)-based tools for diagnostic screening can automate blood smear analysis without relying on a trained technician. Convolutional neural networks (CNN), deep learning neural networks that can identify visual patterns, are being explored for use in abnormality detection in medical images. A parameter that can be optimized in CNN models is the batch size or the number of images used during model training at once in one forward and backward pass. The choice of batch size in developing CNN-based malaria screening tools can affect model accuracy, training speed, and, ultimately, clinical usability. This study explores the impact of batch size on CNN model accuracy for malaria detection from thin blood smear images. Methods We used the publicly available "NIH-NLM-ThinBloodSmearsPf" dataset from the United States National Library of Medicine, consisting of blood smear images for Plasmodium falciparum. The collection consists of 13,779 "parasitized" and 13,779 "uninfected" single-cell images. We created four datasets containing all images, each with unique randomized subsets of images for model testing. Using Python, four identical 10-layer CNN models were developed and trained with varying batch sizes for 10 epochs against all datasets, resulting in 16 sets of outputs. Model prediction accuracy, training time, and F1-score, an accuracy metric used to quantify model performance, were collected. Results All models produced F1-scores of 94%-96%, with 10 of 16 instances producing F1-scores of 95%. After averaging all four dataset outputs by batch size, we observed that, as batch size increased from 16 to 128, the average combined false positives plus false negatives increased by 15.4% (130-150), and the average model F1-score accuracy decreased by 1% (95.3%-94.3%). The average training time also decreased by 28.11% (1,556-1,119 seconds). Conclusion In each dataset, we observe an approximately 1% decrease in F1-score as the batch size was increased. Clinically, a 1% deviation at the population level can create a relatively significant impact on outcomes. Results from this study suggest that smaller batch sizes could improve accuracy in models with similar layer complexity and datasets, potentially resulting in better clinical outcomes. Reduced memory requirement for training also means that model training can be achieved with more economical hardware. Our findings suggest that smaller batch sizes could be evaluated for improvements in accuracy to help develop an AI model that could screen thin blood smears for malaria.
Keywords: artificial intelligence (ai); healthcare technology; malaria screening; medical innovation; public health.
Copyright © 2024, Muralidhar et al.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Automatic patient-level recognition of four Plasmodium species on thin blood smear by a real-time detection transformer (RT-DETR) object detection algorithm: a proof-of-concept and evaluation.Microbiol Spectr. 2024 Feb 6;12(2):e0144023. doi: 10.1128/spectrum.01440-23. Epub 2024 Jan 3. Microbiol Spectr. 2024. PMID: 38171008 Free PMC article.
-
Malaria parasite detection in thick blood smear microscopic images using modified YOLOV3 and YOLOV4 models.BMC Bioinformatics. 2021 Mar 8;22(1):112. doi: 10.1186/s12859-021-04036-4. BMC Bioinformatics. 2021. PMID: 33685401 Free PMC article.
-
iMAGING: a novel automated system for malaria diagnosis by using artificial intelligence tools and a universal low-cost robotized microscope.Front Microbiol. 2023 Nov 24;14:1240936. doi: 10.3389/fmicb.2023.1240936. eCollection 2023. Front Microbiol. 2023. PMID: 38075929 Free PMC article.
-
Development and integration of VGG and dense transfer-learning systems supported with diverse lung images for discovery of the Coronavirus identity.Inform Med Unlocked. 2022;32:101004. doi: 10.1016/j.imu.2022.101004. Epub 2022 Jul 8. Inform Med Unlocked. 2022. PMID: 35822170 Free PMC article. Review.
-
[Artificial intelligence (AI) and hematological diseases: establishment of a peripheral blood convolutional neural network (CNN)-based digital morphology analysis system].Rinsho Ketsueki. 2020;61(5):564-569. doi: 10.11406/rinketsu.61.564. Rinsho Ketsueki. 2020. PMID: 32507825 Review. Japanese.
References
-
- Geneva: World Health Organization. World malaria report 2022. World Malaria Report 2022. [ Apr; 2024 ]. 2022. https://www.who.int/teams/global-malaria-programme/reports/world-malaria... https://www.who.int/teams/global-malaria-programme/reports/world-malaria...
-
- World malaria report 2021. [ Apr; 2024 ];Geneva: World Health Organization. https://www.who.int/teams/global-malaria-programme/reports/world-malaria... World Malaria Report 2021. 2021 1:1.
-
- Malaria. Garcia LS. Clin Lab Med. 2010;30:93–129. - PubMed
LinkOut - more resources
Full Text Sources
