QCMI: A method for quantifying putative biotic associations of microbes at the community level
- PMID: 38868428
- PMCID: PMC10989849
- DOI: 10.1002/imt2.92
QCMI: A method for quantifying putative biotic associations of microbes at the community level
Abstract
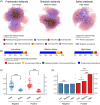
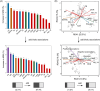
A workflow has been compiled as "qcmi" R package-the quantifying community-level microbial interactions-to identify and quantify the putative biotic associations of microbes at the community level from ecological networks.
© 2023 The Authors. iMeta published by John Wiley & Sons Australia, Ltd on behalf of iMeta Science.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Uncertainty quantification of the effects of biotic interactions on community dynamics from nonlinear time-series data.J R Soc Interface. 2018 Oct 31;15(147):20180695. doi: 10.1098/rsif.2018.0695. J R Soc Interface. 2018. PMID: 30381342 Free PMC article.
-
Diatoms Are Selective Segregators in Global Ocean Planktonic Communities.mSystems. 2020 Jan 21;5(1):e00444-19. doi: 10.1128/mSystems.00444-19. mSystems. 2020. PMID: 31964765 Free PMC article.
-
Applying population and community ecology theory to advance understanding of belowground biogeochemistry.Ecol Lett. 2017 Feb;20(2):231-245. doi: 10.1111/ele.12712. Ecol Lett. 2017. PMID: 28111899 Review.
-
Rhizosphere Soil Microbial Community Under Ice in a High-Latitude Wetland: Different Community Assembly Processes Shape Patterns of Rare and Abundant Microbes.Front Microbiol. 2022 May 23;13:783371. doi: 10.3389/fmicb.2022.783371. eCollection 2022. Front Microbiol. 2022. PMID: 35677902 Free PMC article.
-
Biotic stress-induced changes in root exudation confer plant stress tolerance by altering rhizospheric microbial community.Front Plant Sci. 2023 Mar 10;14:1132824. doi: 10.3389/fpls.2023.1132824. eCollection 2023. Front Plant Sci. 2023. PMID: 36968415 Free PMC article. Review.
Cited by
-
mbDriver: identifying driver microbes in microbial communities based on time-series microbiome data.Brief Bioinform. 2024 Sep 23;25(6):bbae580. doi: 10.1093/bib/bbae580. Brief Bioinform. 2024. PMID: 39526854 Free PMC article.
-
Arbuscular mycorrhizal fungal interactions bridge the support of root-associated microbiota for slope multifunctionality in an erosion-prone ecosystem.Imeta. 2024 Mar 25;3(3):e187. doi: 10.1002/imt2.187. eCollection 2024 Jun. Imeta. 2024. PMID: 38898982 Free PMC article.
References
-
- Delgado‐Baquerizo, Manuel , Reich Peter B., Trivedi Chanda, Eldridge David J., Abades Sebastián, Alfaro Fernando D., Bastida Felip, et al. 2020. “Multiple Elements of Soil Biodiversity Drive Ecosystem Functions Across Biomes.” Nature Ecology & Evolution 4: 210–20. 10.1038/s41559-019-1084-y - DOI - PubMed
LinkOut - more resources
Full Text Sources
