An overview of host-derived molecules that interact with gut microbiota
- PMID: 38868433
- PMCID: PMC10989792
- DOI: 10.1002/imt2.88
An overview of host-derived molecules that interact with gut microbiota
Abstract
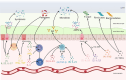
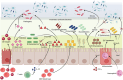
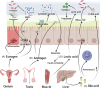
The gut microbiota comprises bacteria, archaea, fungi, protists, and viruses that live together and interact with each other and with host cells. A stable gut microbiota is vital for regulating host metabolism and maintaining body health, while a disturbed microbiota may induce different kinds of disease. In addition, diet is also considered to be the main factor that influences the gut microbiota. The host could shape the gut microbiota through other factors. Here, we reviewed the mechanisms that mediate host regulation on gut microbiota, involved in gut-derived molecules, including gut-derived immune system molecules (secretory immunoglobulin A, antimicrobial peptides, cytokines, cluster of differentiation 4+ effector T cell, and innate lymphoid cells), sources related to gut-derived mucosal molecules (carbon sources, nitrogen sources, oxygen sources, and electron respiratory acceptors), gut-derived exosomal noncoding RNA (ncRNAs) (microRNAs, circular RNA, and long ncRNA), and molecules derived from organs other than the gut (estrogen, androgen, neurohormones, bile acid, and lactic acid). This study provides a systemic overview for understanding the interplay between gut microbiota and host, a comprehensive source for potential ways to manipulate gut microbiota, and a solid foundation for future personalized treatment that utilizes gut microbiota.
Keywords: exosomal ncRNA; gut mucosal molecules; gut‐derived immune molecules; hormones; host; microbiota shaping; molecules from other organs than gut.
© 2023 The Authors. iMeta published by John Wiley & Sons Australia, Ltd on behalf of iMeta Science.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
LinkOut - more resources
Full Text Sources
