Recent advances in the characterization of essential genes and development of a database of essential genes
- PMID: 38868518
- PMCID: PMC10989110
- DOI: 10.1002/imt2.157
Recent advances in the characterization of essential genes and development of a database of essential genes
Abstract
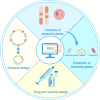
Over the past few decades, there has been a significant interest in the study of essential genes, which are crucial for the survival of an organism under specific environmental conditions and thus have practical applications in the fields of synthetic biology and medicine. An increasing amount of experimental data on essential genes has been obtained with the continuous development of technological methods. Meanwhile, various computational prediction methods, related databases and web servers have emerged accordingly. To facilitate the study of essential genes, we have established a database of essential genes (DEG), which has become popular with continuous updates to facilitate essential gene feature analysis and prediction, drug and vaccine development, as well as artificial genome design and construction. In this article, we summarized the studies of essential genes, overviewed the relevant databases, and discussed their practical applications. Furthermore, we provided an overview of the main applications of DEG and conducted comprehensive analyses based on its latest version. However, it should be noted that the essential gene is a dynamic concept instead of a binary one, which presents both opportunities and challenges for their future development.
Keywords: database of essential genes; drug and vaccine design; essential gene; gene essentiality prediction; genome design.
© 2024 The Authors. iMeta published by John Wiley & Sons Australia, Ltd on behalf of iMeta Science.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
A Comprehensive Overview of Online Resources to Identify and Predict Bacterial Essential Genes.Front Microbiol. 2017 Nov 27;8:2331. doi: 10.3389/fmicb.2017.02331. eCollection 2017. Front Microbiol. 2017. PMID: 29230204 Free PMC article. Review.
-
DEG 5.0, a database of essential genes in both prokaryotes and eukaryotes.Nucleic Acids Res. 2009 Jan;37(Database issue):D455-8. doi: 10.1093/nar/gkn858. Epub 2008 Oct 30. Nucleic Acids Res. 2009. PMID: 18974178 Free PMC article.
-
Machine learning approach to gene essentiality prediction: a review.Brief Bioinform. 2021 Sep 2;22(5):bbab128. doi: 10.1093/bib/bbab128. Brief Bioinform. 2021. PMID: 33842944 Review.
-
Gene essentiality analysis based on DEG, a database of essential genes.Methods Mol Biol. 2008;416:391-400. doi: 10.1007/978-1-59745-321-9_27. Methods Mol Biol. 2008. PMID: 18392983
-
Enhancement of Plant Productivity in the Post-Genomics Era.Curr Genomics. 2016 Aug;17(4):295-6. doi: 10.2174/138920291704160607182507. Curr Genomics. 2016. PMID: 27499678 Free PMC article.
Cited by
-
Systematic identification of cancer-type-specific drugs based on essential genes and validations in lung adenocarcinoma.Brief Bioinform. 2025 May 1;26(3):bbaf266. doi: 10.1093/bib/bbaf266. Brief Bioinform. 2025. PMID: 40483547 Free PMC article.
-
The Roles of T cells in Immune Checkpoint Inhibitor-Induced Arthritis.Aging Dis. 2024 Jul 26;16(4):2100-2119. doi: 10.14336/AD.2024.0546. Aging Dis. 2024. PMID: 39122457 Free PMC article. Review.
-
Application value of nucleic acid MALDI-TOF MS in mycobacterial species identification and drug resistance detection in Mycobacterium tuberculosis.Microbiol Spectr. 2025 Mar 25;13(5):e0154524. doi: 10.1128/spectrum.01545-24. Online ahead of print. Microbiol Spectr. 2025. PMID: 40131854 Free PMC article.
-
Deciphering Rickettsia conorii metabolic pathways: A treasure map to therapeutic targets.Biotechnol Notes. 2024 Nov 23;6:1-9. doi: 10.1016/j.biotno.2024.11.006. eCollection 2025. Biotechnol Notes. 2024. PMID: 39722831 Free PMC article.
-
Genome Analysis of a Newly Sequenced B. subtilis SRCM117797 and Multiple Public B. subtilis Genomes Unveils Insights into Strain Diversification and Biased Core Gene Distribution.Curr Microbiol. 2024 Aug 12;81(10):305. doi: 10.1007/s00284-024-03819-1. Curr Microbiol. 2024. PMID: 39133322
References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous
