Majorbio Cloud: A one-stop, comprehensive bioinformatic platform for multiomics analyses
- PMID: 38868573
- PMCID: PMC10989754
- DOI: 10.1002/imt2.12
Majorbio Cloud: A one-stop, comprehensive bioinformatic platform for multiomics analyses
Abstract
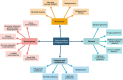
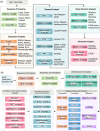
The platform consists of three modules, which are pre-configured bioinformatic pipelines, cloud toolsets, and online omics' courses. The pre-configured bioinformatic pipelines not only combine analytic tools for metagenomics, genomes, transcriptome, proteomics and metabolomics, but also provide users with powerful and convenient interactive analysis reports, which allow them to analyze and mine data independently. As a useful supplement to the bioinformatics pipelines, a wide range of cloud toolsets can further meet the needs of users for daily biological data processing, statistics, and visualization. The rich online courses of multi-omics also provide a state-of-art platform to researchers in interactive communication and knowledge sharing.
© 2022 The Authors. iMeta published by John Wiley & Sons Australia, Ltd on behalf of iMeta Science.
Conflict of interest statement
The platform described in this manuscript is related to an authorized patent CN201810796979.6. Quan Guo, Guo Yu, Yi Ren, Lei Zhang, Yong Zhou, Xianglin Zhang, and Huasheng Huang are inventors of the patent. Huasheng Huang and Xianglin Zhang are cofounders of Majorbio. The other authors are employees of Majorbio.
Figures

References
-
- Keegan, Kevin P. , Glass Elizabeth M., and Meyer Folker. 2016. “MG‐RAST, a Metagenomics Service for Analysis of Microbial Community Structure and Function.” In Microbial Environmental Genomics (MEG), edited by Martin Francis and Uroz Stephane, 207–33. New York: Springer. 10.1007/978-1-4939-3369-3_13 - DOI - PubMed
-
- Jolley, Keith A. , Bray James E., and Maiden Martin C. J.. 2018. “Open‐Access Bacterial Population Genomics: BIGSdb Software, the PubMLST.org Website and their Applications.” Wellcome Open Research 3: 124. 10.12688/wellcomeopenres.14826.1 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
