Generating universal anti-CD19 CAR T cells with a defined memory phenotype by CRISPR/Cas9 editing and safety evaluation of the transcriptome
- PMID: 38868778
- PMCID: PMC11167079
- DOI: 10.3389/fimmu.2024.1401683
Generating universal anti-CD19 CAR T cells with a defined memory phenotype by CRISPR/Cas9 editing and safety evaluation of the transcriptome
Abstract
Introduction: Chimeric antigen receptor-expressing T cells (CAR T cells) have revolutionized cancer treatment, particularly in B cell malignancies. However, the use of autologous T cells for CAR T therapy presents several limitations, including high costs, variable efficacy, and adverse effects linked to cell phenotype.
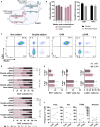
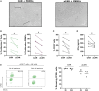
Methods: To overcome these challenges, we developed a strategy to generate universal and safe anti-CD19 CAR T cells with a defined memory phenotype. Our approach utilizes CRISPR/Cas9 technology to target and eliminate the B2M and TRAC genes, reducing graft-versus-host and host-versus-graft responses. Additionally, we selected less differentiated T cells to improve the stability and persistence of the universal CAR T cells. The safety of this method was assessed using our CRISPRroots transcriptome analysis pipeline, which ensures successful gene knockout and the absence of unintended off-target effects on gene expression or transcriptome sequence.
Results: In vitro experiments demonstrated the successful generation of functional universal CAR T cells. These cells exhibited potent lytic activity against tumor cells and a reduced cytokine secretion profile. The CRISPRroots analysis confirmed effective gene knockout and no unintended off-target effects, validating it as a pioneering tool for on/off-target and transcriptome analysis in genome editing experiments.
Discussion: Our findings establish a robust pipeline for manufacturing safe, universal CAR T cells with a favorable memory phenotype. This approach has the potential to address the current limitations of autologous CAR T cell therapy, offering a more stable and persistent treatment option with reduced adverse effects. The use of CRISPRroots enhances the reliability and safety of gene editing in the development of CAR T cell therapies.
Conclusion: We have developed a potent and reliable method for producing universal CAR T cells with a defined memory phenotype, demonstrating both efficacy and safety in vitro. This innovative approach could significantly improve the therapeutic landscape for patients with B cell malignancies.
Keywords: CRISPR/Cas9; CRISPRroots; allogeneic CAR-T cells; anti CD 19 CAR-T cells; memory CAR-T cells.
Copyright © 2024 Pavlovic, Carmona-Luque, Corsi, Maldonado-Pérez, Molina-Estevez, Peralbo-Santaella, Cortijo-Gutiérrez, Justicia-Lirio, Tristán-Manzano, Ronco-Díaz, Ballesteros-Ribelles, Millán-López, Heredia-Velázquez, Fuster-García, Cathomen, Seemann, Gorodkin, Martin, Herrera and Benabdellah.
Conflict of interest statement
Authors MTM and PJL, were employed by LentiStem Biotech. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

