The maternal protein NLRP5 stabilizes UHRF1 in the cytoplasm: implication for the pathogenesis of multilocus imprinting disturbance
- PMID: 38868925
- PMCID: PMC11373322
- DOI: 10.1093/hmg/ddae096
The maternal protein NLRP5 stabilizes UHRF1 in the cytoplasm: implication for the pathogenesis of multilocus imprinting disturbance
Abstract
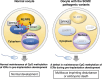
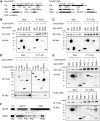
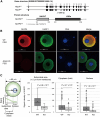
We have recently discovered that the so-called subcortical maternal complex (SCMC) proteins composing of cytoplasmic lattices are destabilized in Uhrf1 knockout murine fully grown oocytes (FGOs). Here we report that human UHRF1 interacts with human NLRP5 and OOEP, which are core components of the SCMC. Moreover, NLRP5 and OOEP interact with DPPA3, which is an essential factor for exporting UHRF1 from the nucleus to the cytoplasm in oocytes. We identify that NLRP5, not OOEP, stabilizes UHRF1 protein in the cytoplasm utilizing specifically engineered cell lines mimicking UHRF1 status in oocytes and preimplantation embryos. Further, UHRF1 is destabilized both in the cytoplasm and nucleus of Nlrp5 knockout murine FGOs. Since pathogenic variants of the SCMC components frequently cause multilocus imprinting disturbance and UHRF1 is essential for maintaining CpG methylation of imprinting control regions during preimplantation development, our results suggest possible pathogenesis behind the disease, which has been a long-standing mystery.
Keywords: MLID; NLRP5; SCMC; UHRF1; multilocus imprinting disturbance.
© The Author(s) 2024. Published by Oxford University Press.
Figures


Similar articles
-
Trans-acting genetic variants causing multilocus imprinting disturbance (MLID): common mechanisms and consequences.Clin Epigenetics. 2022 Mar 16;14(1):41. doi: 10.1186/s13148-022-01259-x. Clin Epigenetics. 2022. PMID: 35296332 Free PMC article.
-
A maternal-effect Padi6 variant causes nuclear and cytoplasmic abnormalities in oocytes, as well as failure of epigenetic reprogramming and zygotic genome activation in embryos.Genes Dev. 2024 Mar 22;38(3-4):131-150. doi: 10.1101/gad.351238.123. Genes Dev. 2024. PMID: 38453481 Free PMC article.
-
Role of UHRF1 in de novo DNA methylation in oocytes and maintenance methylation in preimplantation embryos.PLoS Genet. 2017 Oct 4;13(10):e1007042. doi: 10.1371/journal.pgen.1007042. eCollection 2017 Oct. PLoS Genet. 2017. PMID: 28976982 Free PMC article.
-
The pivotal roles of the NOD-like receptors with a PYD domain, NLRPs, in oocytes and early embryo development†.Biol Reprod. 2019 Aug 1;101(2):284-296. doi: 10.1093/biolre/ioz098. Biol Reprod. 2019. PMID: 31201414 Review.
-
NLRPs, the subcortical maternal complex and genomic imprinting.Reproduction. 2017 Dec;154(6):R161-R170. doi: 10.1530/REP-17-0465. Epub 2017 Sep 15. Reproduction. 2017. PMID: 28916717 Review.
Cited by
-
A Maternal Loss-of-Function Variant in KHDC3L Gene Causes a Range of Adverse Pregnancy Outcomes: A Case Report.Mol Genet Genomic Med. 2025 Jan;13(1):e70051. doi: 10.1002/mgg3.70051. Mol Genet Genomic Med. 2025. PMID: 39763182 Free PMC article.
-
The subcortical maternal complex safeguards mouse oocyte-to-embryo transition by preventing nuclear entry of SPIN1.Nat Struct Mol Biol. 2025 Aug;32(8):1358-1372. doi: 10.1038/s41594-025-01538-0. Epub 2025 Apr 17. Nat Struct Mol Biol. 2025. PMID: 40247146
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

