Dissecting Acute Drug-Induced Hepatotoxicity and Therapeutic Responses of Steatotic Liver Disease Using Primary Mouse Liver and Blood Cells in a Liver-On-A-Chip Model
- PMID: 38868948
- PMCID: PMC11321671
- DOI: 10.1002/advs.202403516
Dissecting Acute Drug-Induced Hepatotoxicity and Therapeutic Responses of Steatotic Liver Disease Using Primary Mouse Liver and Blood Cells in a Liver-On-A-Chip Model
Abstract
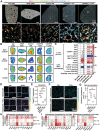
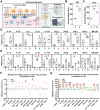
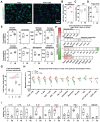
Metabolic dysfunction-associated steatotic liver disease (MASLD) is hallmarked by hepatic steatosis, cell injury, inflammation, and fibrosis. This study elaborates on a multicellular biochip-based liver sinusoid model to mimic MASLD pathomechanisms and investigate the therapeutic effects of drug candidates lanifibranor and resmetirom. Mouse liver primary hepatocytes, hepatic stellate cells, Kupffer cells, and endothelial cells are seeded in a dual-chamber biocompatible liver-on-a-chip (LoC). The LoC is then perfused with circulating immune cells (CICs). Acetaminophen (APAP) and free fatty acids (FFAs) treatment recapitulate acute drug-induced liver injury and MASLD, respectively. As a benchmark for the LoC, multiplex immunofluorescence on livers from APAP-injected and dietary MASLD-induced mice reveals characteristic changes on parenchymal and immune cell populations. APAP exposure induces cell death in the LoC, and increased inflammatory cytokine levels in the circulating perfusate. Under FFA stimulation, lipid accumulation, cellular damage, inflammatory secretome, and fibrogenesis are increased in the LoC, reflecting MASLD. Both injury conditions potentiate CIC migration from the perfusate to the LoC cellular layers. Lanifibranor prevents the onset of inflammation, while resmetirom decreases lipid accumulation in hepatocytes and increases the generation of FFA metabolites in the LoC. This study demonstrates the LoC potential for functional and molecular evaluation of liver disease drug candidates.
Keywords: NAFLD; NASH; fibrosis; liver diseases; macrophages; microfluidic biochip; steatohepatitis.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
A.S.M. holds equity in and consults for Dynamic42 GmbH. FT's lab has received research funding from Allergan, Bristol‐Myers Squibb, Gilead and Inventiva. FT has received honoraria for consulting or lectures from Astra Zeneca, Gilead, AbbVie, BMS, Boehringer, Madrigal, MSD, GSK, Intercept, Falk, Ionis, Inventiva, Merz, Pfizer, Alnylam, NGM, CSL Behring, Novo Nordisk, Novartis. The other authors declare no conflict of interest.
Figures



References
-
- Lazarus J. V., Mark H. E., Allen A. M., Arab J. P., Carrieri P., Noureddin M., Alazawi W., Alkhouri N., Alqahtani S. A., Anstee Q. M., Arrese M., Bataller R., Berg T., Brennan P. N., Burra P., Castro‐Narro G. E., Cortez‐Pinto H., Cusi K., Dedes N., Duseja A., Francque S. M., Gastaldelli A., Hagstrom H., Huang T. T. K., Ivancovsky Wajcman D., Kautz A., Kopka C. J., Krag A., Newsome P. N., Rinella M. E., et al., Hepatology. 2023, 79, 502. - PMC - PubMed
-
- Rinella M. E., Lazarus J. V., Ratziu V., Francque S. M., Sanyal A. J., Kanwal F., Romero D., Abdelmalek M. F., Anstee Q. M., Arab J. P., Arrese M., Bataller R., Beuers U., Boursier J., Bugianesi E., Byrne C., Castro Narro G. E., Chowdhury A., Cortez‐Pinto H., Cryer D., Cusi K., El‐Kassas M., Klein S., Eskridge W., Fan J., Gawrieh S., Guy C. D., Harrison S. A., Kim S. U., Koot B., et al., J Hepatol. 2023, 79, 1542. - PubMed
-
- Mitten E. K., Baffy G., J. Hepatol. 2022, 77, 1642. - PubMed
-
- Powell E. E., Wong V. W., Rinella M., Lancet. 2021, 397, 2212. - PubMed
-
- Tacke F., Puengel T., Loomba R., Friedman S. L., J. Hepatol. 2023, 79, 552. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
