CCDC50 mediates the clearance of protein aggregates to prevent cellular proteotoxicity
- PMID: 38869076
- PMCID: PMC11572255
- DOI: 10.1080/15548627.2024.2367183
CCDC50 mediates the clearance of protein aggregates to prevent cellular proteotoxicity
Abstract
Protein aggregation caused by the disruption of proteostasis will lead to cellular cytotoxicity and even cell death, which is implicated in multiple neurodegenerative diseases. The elimination of aggregated proteins is mediated by selective macroautophagy receptors, which is termed aggrephagy. However, the identity and redundancy of aggrephagy receptors in recognizing substrates remain largely unexplored. Here, we find that CCDC50, a highly expressed autophagy receptor in brain, is recruited to proteotoxic stresses-induced polyubiquitinated protein aggregates and ectopically expressed aggregation-prone proteins. CCDC50 recognizes and further clears these cytotoxic aggregates through autophagy. The ectopic expression of CCDC50 increases the tolerance to stress-induced proteotoxicity and hence improved cell survival in neuron cells, whereas CCDC50 deficiency caused accumulation of lipid deposits and polyubiquitinated protein conjugates in the brain of one-year-old mice. Our study illustrates how aggrephagy receptor CCDC50 combats proteotoxic stress for the benefit of neuronal cell survival, thus suggesting a protective role in neurotoxic proteinopathy.Abbreviations: AD: Alzheimer disease; ALS: amyotrophic lateral sclerosis; ATG5: autophagy related 5; BODIPY: boron-dipyrromethene; CASP3: caspase 3; CCDC50: coiled-coil domain containing 50; CCT2: chaperonin containing TCP1 subunit 2; CHX: cycloheximide; CQ: chloroquine; CRISPR: clustered regulatory interspaced short palindromic repeat; Cas9: CRISPR-associated system 9; DAPI: 4',6-diamidino-2-phenylindole; FK2: Anti-ubiquitinylated proteins antibody, clone FK2; FUS: FUS RNA binding protein; GFP: green fluorescent protein; HD: Huntington disease; HTT: huntingtin; KEGG: Kyoto Encyclopedia of Genes and Genomes; LDS: LIR-docking site; LIR: LC3-interacting region; MAP1LC3/LC3: microtubule associated protein 1 light chain 3; MAPT/tau: microtubule associated protein tau; MIU: motif interacting with ubiquitin; NBR1: NBR1, autophagy cargo receptor; OPTN: optineurin; PD: Parkinson disease; PI: propidium iodide; ROS: reactive oxygen species; SOD1: superoxide dismutase 1; SQSTM1/p62: sequestosome 1; TAX1BP1: Tax1 binding protein 1; Ub: ubiquitin; UDS: UIM-docking site; UIM: ubiquitin interacting motif; UPS: ubiquitin-proteasome system.
Keywords: Aggrephagy receptor; CCDC50; neurodegenerative diseases; protein aggregation; proteostasis.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures
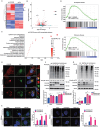
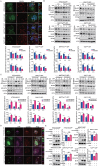
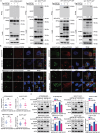
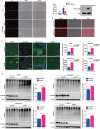
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
