Multi-transcriptomics identifies targets of the endoribonuclease DNE1 and highlights its coordination with decapping
- PMID: 38869231
- PMCID: PMC11371186
- DOI: 10.1093/plcell/koae175
Multi-transcriptomics identifies targets of the endoribonuclease DNE1 and highlights its coordination with decapping
Abstract
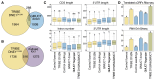
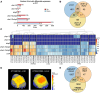
Decapping is a crucial step in mRNA degradation in eucaryotes and requires the formation of a holoenzyme complex between the decapping enzyme DECAPPING 2 (DCP2) and the decapping enhancer DCP1. In Arabidopsis (Arabidopsis thaliana), DCP1-ASSOCIATED NYN ENDORIBONUCLEASE 1 (DNE1) is a direct protein partner of DCP1. The function of both DNE1 and decapping is necessary to maintain phyllotaxis, the regularity of organ emergence in the apex. In this study, we combined in vivo mRNA editing, RNA degradome sequencing, transcriptomics, and small RNA-omics to identify targets of DNE1 and study how DNE1 and DCP2 cooperate in controlling mRNA fate. Our data reveal that DNE1 mainly contacts and cleaves mRNAs in the coding sequence and has sequence cleavage preferences. DNE1 targets are also degraded through decapping, and both RNA degradation pathways influence the production of mRNA-derived small interfering RNAs. Finally, we detected mRNA features enriched in DNE1 targets including RNA G-quadruplexes and translated upstream open reading frames. Combining these four complementary high-throughput sequencing strategies greatly expands the range of DNE1 targets and allowed us to build a conceptual framework describing the influence of DNE1 and decapping on mRNA fate. These data will be crucial to unveil the specificity of DNE1 action and understand its importance for developmental patterning.
© The Author(s) 2024. Published by Oxford University Press on behalf of American Society of Plant Biologists. All rights reserved. For commercial re-use, please contact reprints@oup.com for reprints and translation rights for reprints. All other permissions can be obtained through our RightsLink service via the Permissions link on the article page on our site—for further information please contact journals.permissions@oup.com.
Conflict of interest statement
Conflict of interest statement. None declared.
Figures





References
-
- Branscheid A, Marchais A, Schott G, Lange H, Gagliardi D, Andersen SU, Voinnet O, Brodersen P. SKI2 mediates degradation of RISC 5′-cleavage fragments and prevents secondary siRNA production from miRNA targets in Arabidopsis. Nucleic Acids Res. 2015:43(22):10975–10988. 10.1093/nar/gkv1014 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

