Gain-of-function human UNC93B1 variants cause systemic lupus erythematosus and chilblain lupus
- PMID: 38869500
- PMCID: PMC11176256
- DOI: 10.1084/jem.20232066
Gain-of-function human UNC93B1 variants cause systemic lupus erythematosus and chilblain lupus
Abstract
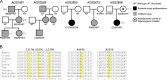
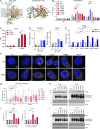
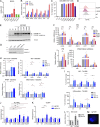
UNC93B1 is a transmembrane domain protein mediating the signaling of endosomal Toll-like receptors (TLRs). We report five families harboring rare missense substitutions (I317M, G325C, L330R, R466S, and R525P) in UNC93B1 causing systemic lupus erythematosus (SLE) or chilblain lupus (CBL) as either autosomal dominant or autosomal recessive traits. As for a D34A mutation causing murine lupus, we recorded a gain of TLR7 and, to a lesser extent, TLR8 activity with the I317M (in vitro) and G325C (in vitro and ex vivo) variants in the context of SLE. Contrastingly, in three families segregating CBL, the L330R, R466S, and R525P variants were isomorphic with respect to TLR7 activity in vitro and, for R525P, ex vivo. Rather, these variants demonstrated a gain of TLR8 activity. We observed enhanced interaction of the G325C, L330R, and R466S variants with TLR8, but not the R525P substitution, indicating different disease mechanisms. Overall, these observations suggest that UNC93B1 mutations cause monogenic SLE or CBL due to differentially enhanced TLR7 and TLR8 signaling.
© 2024 David et al.
Conflict of interest statement
Disclosures: C. Paul reported personal fees from Abbvie, Boehringer, BMS, Eli Lilly, and Janssen; grants from Novartis; and personal fees from Pfizer, Sanofi, Pierre Fabre, and UCB outside the submitted work. D. Bonnet reported personal fees from Novartis and MSD outside the submitted work. No other disclosures were reported.
Figures




References
-
- Al-Azab, M., Idiiatullina E., Lin M., Hrovat-Schaale K., Xian H., Zhu J., Yang M., Lu B., Liu Z., Zhao Z., et al. . 2023. Genetic variants in UNC93B1 predispose to childhood-onset systemic lupus erythematosus. medRxiv. 10.1101/2023.10.15.23296507 (Preprint posted October 16, 2023). - DOI - PMC - PubMed
-
- Aluri, J., Bach A., Kaviany S., Chiquetto Paracatu L., Kitcharoensakkul M., Walkiewicz M.A., Putnam C.D., Shinawi M., Saucier N., Rizzi E.M., et al. . 2021. Immunodeficiency and bone marrow failure with mosaic and germline TLR8 gain of function. Blood. 137:2450–2462. 10.1182/blood.2020009620 - DOI - PMC - PubMed
-
- Asano, T., Boisson B., Onodi F., Matuozzo D., Moncada-Velez M., Maglorius Renkilaraj M.R.L., Zhang P., Meertens L., Bolze A., Materna M., et al. . 2021. X-linked recessive TLR7 deficiency in ∼1% of men under 60 years old with life-threatening COVID-19. Sci. Immunol. 6:eabl4348. 10.1126/sciimmunol.abl4348 - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
- FDM202106013329/Fondation pour la Recherche Médicale
- 786142 E-T1IFNs/ERC_/European Research Council/International
- MC_UU_00035/11/MRC_/Medical Research Council/United Kingdom
- ANR-10-IAHU-01/Agence Nationale de la Recherche
- HHMI/Howard Hughes Medical Institute/United States
- Rockefeller University
- St. Giles Foundation
- EQU201903007798/French Foundation for Medical Research
- 824110/European Union's Horizon 2020 research and innovation program
- Square Foundation
- Grandir-Fonds de solidarité pour l'enfance
- General Atlantic Foundation
- Institut National de la Santé et de la Recherche Médicale
- Paris Cité University
- European Molecular Biology Organization
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

