C16ORF70/MYTHO promotes healthy aging in C.elegans and prevents cellular senescence in mammals
- PMID: 38869949
- PMCID: PMC11291266
- DOI: 10.1172/JCI165814
C16ORF70/MYTHO promotes healthy aging in C.elegans and prevents cellular senescence in mammals
Abstract
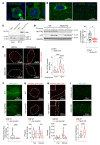
The identification of genes that confer either extension of life span or accelerate age-related decline was a step forward in understanding the mechanisms of aging and revealed that it is partially controlled by genetics and transcriptional programs. Here, we discovered that the human DNA sequence C16ORF70 encodes a protein, named MYTHO (macroautophagy and youth optimizer), which controls life span and health span. MYTHO protein is conserved from Caenorhabditis elegans to humans and its mRNA was upregulated in aged mice and elderly people. Deletion of the orthologous myt-1 gene in C. elegans dramatically shortened life span and decreased animal survival upon exposure to oxidative stress. Mechanistically, MYTHO is required for autophagy likely because it acts as a scaffold that binds WIPI2 and BCAS3 to recruit and assemble the conjugation system at the phagophore, the nascent autophagosome. We conclude that MYTHO is a transcriptionally regulated initiator of autophagy that is central in promoting stress resistance and healthy aging.
Keywords: Aging; Autophagy; Cell biology; Cellular senescence; Skeletal muscle.
Figures








References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

