SIRT7 promotes lung cancer progression by destabilizing the tumor suppressor ARF
- PMID: 38870055
- PMCID: PMC11194565
- DOI: 10.1073/pnas.2409269121
SIRT7 promotes lung cancer progression by destabilizing the tumor suppressor ARF
Abstract
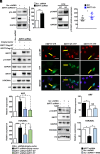
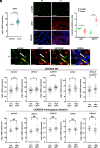
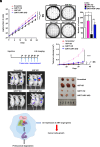
Sirtuin 7 (SIRT7) is a member of the mammalian family of nicotinamide adenine dinucleotide (NAD+)-dependent histone/protein deacetylases, known as sirtuins. It acts as a potent oncogene in numerous malignancies, but the molecular mechanisms employed by SIRT7 to sustain lung cancer progression remain largely uncharacterized. We demonstrate that SIRT7 exerts oncogenic functions in lung cancer cells by destabilizing the tumor suppressor alternative reading frame (ARF). SIRT7 directly interacts with ARF and prevents binding of ARF to nucleophosmin, thereby promoting proteasomal-dependent degradation of ARF. We show that SIRT7-mediated degradation of ARF increases expression of protumorigenic genes and stimulates proliferation of non-small-cell lung cancer (NSCLC) cells both in vitro and in vivo in a mouse xenograft model. Bioinformatics analysis of transcriptome data from human lung adenocarcinomas revealed a correlation between SIRT7 expression and increased activity of genes normally repressed by ARF. We propose that disruption of SIRT7-ARF signaling stabilizes ARF and thus attenuates cancer cell proliferation, offering a strategy to mitigate NSCLC progression.
Keywords: ARF; SIRT7; Sirtuins; lung cancer; nucleophosmin.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures




References
-
- Kim W. Y., Sharpless N. E., The regulation of INK4/ARF in cancer and aging. Cell 127, 265–275 (2006). - PubMed
-
- Kamijo T., et al. , Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell 91, 649–659 (1997). - PubMed
-
- Serrano M., et al. , Role of the INK4a locus in tumor suppression and cell mortality. Cell 85, 27–37 (1996). - PubMed
-
- Sherr C. J., Divorcing ARF and p53: An unsettled case. Nat. Rev. Cancer 6, 663–673 (2006). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

