The multifunction Coxiella effector Vice stimulates macropinocytosis and interferes with the ESCRT machinery
- PMID: 38870060
- PMCID: PMC11194487
- DOI: 10.1073/pnas.2315481121
The multifunction Coxiella effector Vice stimulates macropinocytosis and interferes with the ESCRT machinery
Abstract
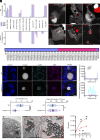
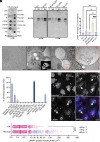
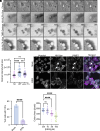
Intracellular bacterial pathogens divert multiple cellular pathways to establish their niche and persist inside their host. Coxiella burnetii, the causative agent of Q fever, secretes bacterial effector proteins via its Type 4 secretion system to generate a Coxiella-containing vacuole (CCV). Manipulation of lipid and protein trafficking by these effectors is essential for bacterial replication and virulence. Here, we have characterized the lipid composition of CCVs and found that the effector Vice interacts with phosphoinositides and membranes enriched in phosphatidylserine and lysobisphosphatidic acid. Remarkably, eukaryotic cells ectopically expressing Vice present compartments that resemble early CCVs in both morphology and composition. We found that the biogenesis of these compartments relies on the double function of Vice. The effector protein initially localizes at the plasma membrane of eukaryotic cells where it triggers the internalization of large vacuoles by macropinocytosis. Then, Vice stabilizes these compartments by perturbing the ESCRT machinery. Collectively, our results reveal that Vice is an essential C. burnetii effector protein capable of hijacking two major cellular pathways to shape the bacterial replicative niche.
Keywords: Coxiella burnetii; ESCRT; effector protein; lipids; macropinocytosis.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures

References
-
- Martinez E., Siadous F. A., Bonazzi M., Tiny architects: Biogenesis of intracellular replicative niches by bacterial pathogens. Fems Microbiol. Rev. 42, 425–447 (2018). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

