Broccoli aptamer allows quantitative transcription regulation studies in vitro
- PMID: 38870160
- PMCID: PMC11175446
- DOI: 10.1371/journal.pone.0304677
Broccoli aptamer allows quantitative transcription regulation studies in vitro
Abstract
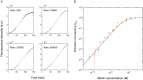
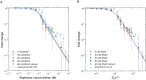
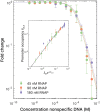
Quantitative transcription regulation studies in vivo and in vitro often make use of reporter proteins. Here we show that using Broccoli aptamers, quantitative study of transcription in various regulatory scenarios is possible without a translational step. To explore the method we studied several regulatory scenarios that we analyzed using thermodynamic occupancy-based models, and found excellent agreement with previous studies. In the next step we show that non-coding DNA can have a dramatic effect on the level of transcription, similar to the influence of the lac repressor with a strong affinity to operator sites. Finally, we point out the limitations of the method in terms of delay times coupled to the folding of the aptamer. We conclude that the Broccoli aptamer is suitable for quantitative transcription measurements.
Copyright: © 2024 van der Sijs et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Revisiting T7 RNA polymerase transcription in vitro with the Broccoli RNA aptamer as a simplified real-time fluorescent reporter.J Biol Chem. 2021 Jan-Jun;296:100175. doi: 10.1074/jbc.RA120.014553. Epub 2020 Dec 16. J Biol Chem. 2021. PMID: 33303627 Free PMC article.
-
Fluorophore-Promoted RNA Folding and Photostability Enables Imaging of Single Broccoli-Tagged mRNAs in Live Mammalian Cells.Angew Chem Int Ed Engl. 2020 Mar 9;59(11):4511-4518. doi: 10.1002/anie.201914576. Epub 2020 Jan 28. Angew Chem Int Ed Engl. 2020. PMID: 31850609 Free PMC article.
-
In-gel imaging of RNA processing using broccoli reveals optimal aptamer expression strategies.Chem Biol. 2015 May 21;22(5):649-60. doi: 10.1016/j.chembiol.2015.04.018. Chem Biol. 2015. PMID: 26000751 Free PMC article.
-
In Vivo Production of RNA Aptamers and Nanoparticles: Problems and Prospects.Molecules. 2021 Mar 6;26(5):1422. doi: 10.3390/molecules26051422. Molecules. 2021. PMID: 33800717 Free PMC article. Review.
-
Methods for Improving Aptamer Binding Affinity.Molecules. 2016 Mar 28;21(4):421. doi: 10.3390/molecules21040421. Molecules. 2016. PMID: 27043498 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

