Computational Analysis of Heat Capacity Effects in Protein-Ligand Binding
- PMID: 38870420
- PMCID: PMC11238534
- DOI: 10.1021/acs.jctc.4c00525
Computational Analysis of Heat Capacity Effects in Protein-Ligand Binding
Abstract
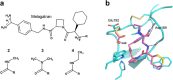
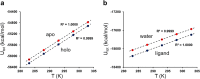
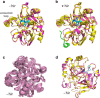
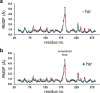
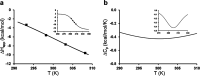
Heat capacity effects in protein-ligand binding as measured by calorimetric experiments have recently attracted considerable attention, particularly in the field of enzyme inhibitor design. A significant negative heat capacity change upon ligand binding implies a marked temperature dependence of the binding enthalpy, which is of high relevance for attempts to optimize protein-ligand interactions. In this work, we address the question of how well such heat capacity changes can be predicted by computer simulations. We examine a series of human thrombin inhibitors that all bind with ΔCp values of about -0.4 kcal/mol/K and calculate heat capacity changes from plain molecular dynamics simulations of the bound and free states of the enzyme and ligand. The results show that accurate ΔCp estimates within a few tenths of a kcal/mol/K of the experimental values can be obtained with this approach. This allows us to address the structural and energetic origin of the negative heat capacity changes for the thrombin inhibitors, and it is found that conformational equilibria of the free ligands in solution make a major contribution to the observed effect.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

Similar articles
-
Thermodynamic investigation of hirudin binding to the slow and fast forms of thrombin: evidence for folding transitions in the inhibitor and protease coupled to binding.J Mol Biol. 1995 Nov 10;253(5):787-98. doi: 10.1006/jmbi.1995.0591. J Mol Biol. 1995. PMID: 7473752
-
Thermodynamics of substrates and reversible inhibitors binding to the active site cleft of human alpha-thrombin.J Mol Biol. 1994 Jun 17;239(4):569-77. doi: 10.1006/jmbi.1994.1396. J Mol Biol. 1994. PMID: 8006969
-
On the link between conformational changes, ligand binding and heat capacity.Biochim Biophys Acta. 2016 May;1860(5):868-878. doi: 10.1016/j.bbagen.2015.10.010. Epub 2015 Oct 22. Biochim Biophys Acta. 2016. PMID: 26476135 Review.
-
Heat Capacity Changes for Transition-State Analogue Binding and Catalysis with Human 5'-Methylthioadenosine Phosphorylase.ACS Chem Biol. 2017 Feb 17;12(2):464-473. doi: 10.1021/acschembio.6b00885. Epub 2016 Dec 27. ACS Chem Biol. 2017. PMID: 28026167 Free PMC article.
-
Thermodynamics of protein-ligand interactions: history, presence, and future aspects.J Recept Signal Transduct Res. 2004 Feb;24(1-2):1-52. doi: 10.1081/rrs-120037896. J Recept Signal Transduct Res. 2004. PMID: 15344878 Review.
Cited by
-
Impact of high temperatures on enzyme-linked immunoassay (ELISA) performance for leptin measurements in human milk stored under varied freeze/thaw conditions.PLoS One. 2025 Mar 19;20(3):e0320366. doi: 10.1371/journal.pone.0320366. eCollection 2025. PLoS One. 2025. PMID: 40106448 Free PMC article.
-
Accurate Prediction of Drug Activity by Computational Methods: Importance of Thermal Capacity.Molecules. 2025 Jun 12;30(12):2563. doi: 10.3390/molecules30122563. Molecules. 2025. PMID: 40572530 Free PMC article.
-
Computer Simulations of the Temperature Dependence of Enzyme Reactions.J Chem Theory Comput. 2025 Feb 11;21(3):1017-1028. doi: 10.1021/acs.jctc.4c01733. Epub 2025 Jan 30. J Chem Theory Comput. 2025. PMID: 39884967 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
