Rescoring Peptide Spectrum Matches: Boosting Proteomics Performance by Integrating Peptide Property Predictors Into Peptide Identification
- PMID: 38871251
- PMCID: PMC11269915
- DOI: 10.1016/j.mcpro.2024.100798
Rescoring Peptide Spectrum Matches: Boosting Proteomics Performance by Integrating Peptide Property Predictors Into Peptide Identification
Abstract
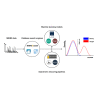
Rescoring of peptide spectrum matches originating from database search engines enabled by peptide property predictors is exceeding the performance of peptide identification from traditional database search engines. In contrast to the peptide spectrum match scores calculated by traditional database search engines, rescoring peptide spectrum matches generates scores based on comparing observed and predicted peptide properties, such as fragment ion intensities and retention times. These newly generated scores enable a more efficient discrimination between correct and incorrect peptide spectrum matches. This approach was shown to lead to substantial improvements in the number of confidently identified peptides, facilitating the analysis of challenging datasets in various fields such as immunopeptidomics, metaproteomics, proteogenomics, and single-cell proteomics. In this review, we summarize the key elements leading up to the recent introduction of multiple data-driven rescoring pipelines. We provide an overview of relevant post-processing rescoring tools, introduce prominent data-driven rescoring pipelines for various applications, and highlight limitations, opportunities, and future perspectives of this approach and its impact on mass spectrometry-based proteomics.
Keywords: artificial intelligence; computational proteomics; data-driven rescoring; machine learning; peptide identification; peptide property prediction; rescoring.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: M. W. is a co-founder and shareholder of MSAID GmbH and OmicScouts GmbH, with no operational role in both companies.
Figures


Similar articles
-
INFERYS rescoring: Boosting peptide identifications and scoring confidence of database search results.Rapid Commun Mass Spectrom. 2025 May;39 Suppl 1:e9128. doi: 10.1002/rcm.9128. Epub 2021 Jun 28. Rapid Commun Mass Spectrom. 2025. PMID: 34015160
-
Machine learning-based peptide-spectrum match rescoring opens up the immunopeptidome.Proteomics. 2024 Apr;24(8):e2300336. doi: 10.1002/pmic.202300336. Epub 2023 Nov 27. Proteomics. 2024. PMID: 38009585 Review.
-
Comparative Analysis of Data-Driven Rescoring Platforms for Improved Peptide Identification in HeLa Digest Samples.Proteomics. 2025 Apr;25(7):e202400225. doi: 10.1002/pmic.202400225. Epub 2025 Feb 2. Proteomics. 2025. PMID: 39895169 Free PMC article.
-
inSPIRE: An Open-Source Tool for Increased Mass Spectrometry Identification Rates Using Prosit Spectral Prediction.Mol Cell Proteomics. 2022 Dec;21(12):100432. doi: 10.1016/j.mcpro.2022.100432. Epub 2022 Oct 21. Mol Cell Proteomics. 2022. PMID: 36280141 Free PMC article.
-
Peptide Property Prediction for Mass Spectrometry Using AI: An Introduction to State of the Art Models.Proteomics. 2025 May;25(9-10):e202400398. doi: 10.1002/pmic.202400398. Epub 2025 Apr 10. Proteomics. 2025. PMID: 40211610 Free PMC article. Review.
Cited by
-
Proteome-wide non-cleavable crosslink identification with MS Annika 3.0 reveals the structure of the C. elegans Box C/D complex.Commun Chem. 2024 Dec 19;7(1):300. doi: 10.1038/s42004-024-01386-x. Commun Chem. 2024. PMID: 39702463 Free PMC article.
-
Prosit-XL: enhanced cross-linked peptide identification by fragment intensity prediction to study protein interactions and structures.Nat Commun. 2025 Jul 1;16(1):5429. doi: 10.1038/s41467-025-61203-4. Nat Commun. 2025. PMID: 40592844 Free PMC article.
-
The 2024 Report on the Human Proteome from the HUPO Human Proteome Project.J Proteome Res. 2024 Dec 6;23(12):5296-5311. doi: 10.1021/acs.jproteome.4c00776. Epub 2024 Nov 8. J Proteome Res. 2024. PMID: 39514846 Free PMC article. Review.
-
Rustims: An Open-Source Framework for Rapid Development and Processing of timsTOF Data-Dependent Acquisition Data.J Proteome Res. 2025 May 2;24(5):2358-2368. doi: 10.1021/acs.jproteome.4c00966. Epub 2025 Apr 22. J Proteome Res. 2025. PMID: 40260647 Free PMC article.
-
MSCI: an open-source Python package for information content assessment of peptide fragmentation spectra.Bioinform Adv. 2025 May 24;5(1):vbaf125. doi: 10.1093/bioadv/vbaf125. eCollection 2025. Bioinform Adv. 2025. PMID: 40584887 Free PMC article.
References
-
- Petrosius V., Aragon-Fernandez P., Arrey T.N., Üresin N., Furtwängler B., Stewart H., et al. Evaluating the capabilities of the Astral mass analyzer for single-cell proteomics. bioRxiv. 2023 doi: 10.1101/2023.06.06.543943. [preprint] - DOI
-
- Zhang H., Ouyang Z., Zhang W. Advances in mass spectrometry for clinical analysis: data acquisition, interpretation and information integration. Trac Trends Anal. Chem. 2023;169
-
- Kresse M., Drinda H., Romanotto A., Speer K. Simultaneous determination of pesticides, mycotoxins, and metabolites as well as other contaminants in cereals by LC-LC-MS/MS. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2019;1117:86–102. - PubMed
-
- Eng J.K., Jahan T.A., Hoopmann M.R. Comet: an open-source MS/MS sequence database search tool. Proteomics. 2013;13:22–24. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

