Genomic characterization of cervical lymph node metastases in papillary thyroid carcinoma following the Chornobyl accident
- PMID: 38871684
- PMCID: PMC11176192
- DOI: 10.1038/s41467-024-49292-z
Genomic characterization of cervical lymph node metastases in papillary thyroid carcinoma following the Chornobyl accident
Abstract
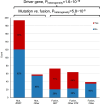
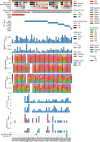
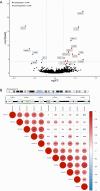
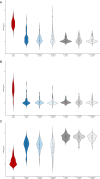
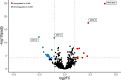
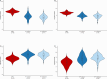
Childhood radioactive iodine exposure from the Chornobyl accident increased papillary thyroid carcinoma (PTC) risk. While cervical lymph node metastases (cLNM) are well-recognized in pediatric PTC, the PTC metastatic process and potential radiation association are poorly understood. Here, we analyze cLNM occurrence among 428 PTC with genomic landscape analyses and known drivers (131I-exposed = 349, unexposed = 79; mean age = 27.9 years). We show that cLNM are more frequent in PTC with fusion (55%) versus mutation (30%) drivers, although the proportion varies by specific driver gene (RET-fusion = 71%, BRAF-mutation = 38%, RAS-mutation = 5%). cLNM frequency is not associated with other characteristics, including radiation dose. cLNM molecular profiling (N = 47) demonstrates 100% driver concordance with matched primary PTCs and highly concordant mutational spectra. Transcriptome analysis reveals 17 differentially expressed genes, particularly in the HOXC cluster and BRINP3; the strongest differentially expressed microRNA also is near HOXC10. Our findings underscore the critical role of driver alterations and provide promising candidates for elucidating the biological underpinnings of PTC cLNM.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
E.T.D. is an employee of Nvidia Corporation and owns stock in Nvidia, Illumina, and Pacific Biosciences. G.G. receives research funds from IBM and Pharmacyclics, and is an inventor on patent applications related to MuTect, ABSOLUTE, MutSig, MSMuTect, MSMutSig, MSIdetect, POLYSOLVER, and TensorQTL. G.G. is a founder, consultant and holds privately held equity in Scorpion Therapeutics. The remaining authors declare no competing interests.
Figures

References
-
- Howlader, N. et al. SEER cancer statistics review, 1975-2018. National Cancer Institute. https://seer.cancer.gov/csr/1975_2018/ (2021).
-
- Haugen BR, et al. 2015 American Thyroid Association Management Guidelines for adult patients with thyroid nodules and differentiated thyroid cancer: The American Thyroid Association Guidelines Task Force on thyroid nodules and differentiated thyroid cancer. Thyroid. 2016;26:1–133. doi: 10.1089/thy.2015.0020. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

