Potential pandemic risk of circulating swine H1N2 influenza viruses
- PMID: 38871701
- PMCID: PMC11176300
- DOI: 10.1038/s41467-024-49117-z
Potential pandemic risk of circulating swine H1N2 influenza viruses
Abstract
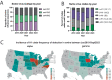
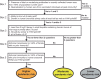
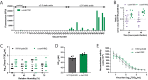
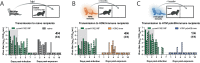
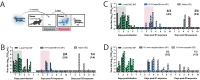
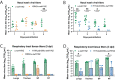
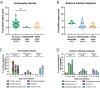
Influenza A viruses in swine have considerable genetic diversity and continue to pose a pandemic threat to humans due to a potential lack of population level immunity. Here we describe a pipeline to characterize and triage influenza viruses for their pandemic risk and examine the pandemic potential of two widespread swine origin viruses. Our analysis reveals that a panel of human sera collected from healthy adults in 2020 has no cross-reactive neutralizing antibodies against a α-H1 clade strain (α-swH1N2) but do against a γ-H1 clade strain. The α-swH1N2 virus replicates efficiently in human airway cultures and exhibits phenotypic signatures similar to the human H1N1 pandemic strain from 2009 (H1N1pdm09). Furthermore, α-swH1N2 is capable of efficient airborne transmission to both naïve ferrets and ferrets with prior seasonal influenza immunity. Ferrets with H1N1pdm09 pre-existing immunity show reduced α-swH1N2 viral shedding and less severe disease signs. Despite this, H1N1pdm09-immune ferrets that became infected via the air can still onward transmit α-swH1N2 with an efficiency of 50%. These results indicate that this α-swH1N2 strain has a higher pandemic potential, but a moderate level of impact since there is reduced replication fitness and pathology in animals with prior immunity.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
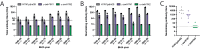
References
MeSH terms
Substances
Grants and funding
- R01 AI158484/AI/NIAID NIH HHS/United States
- CAMS 1013362.02/Burroughs Wellcome Fund (BWF)
- T32 AI138954/AI/NIAID NIH HHS/United States
- 75N93021C00015/AI/NIAID NIH HHS/United States
- 5030-32000-231-000-D/United States Department of Agriculture | Agricultural Research Service (USDA Agricultural Research Service)
LinkOut - more resources
Full Text Sources

