A non-methanogenic archaeon within the order Methanocellales
- PMID: 38871712
- PMCID: PMC11176372
- DOI: 10.1038/s41467-024-48185-5
A non-methanogenic archaeon within the order Methanocellales
Abstract
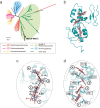
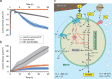
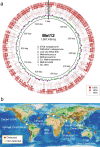
Serpentinization, a geochemical process found on modern and ancient Earth, provides an ultra-reducing environment that can support microbial methanogenesis and acetogenesis. Several groups of archaea, such as the order Methanocellales, are characterized by their ability to produce methane. Here, we generate metagenomic sequences from serpentinized springs in The Cedars, California, and construct a circularized metagenome-assembled genome of a Methanocellales archaeon, termed Met12, that lacks essential methanogenesis genes. The genome includes genes for an acetyl-CoA pathway, but lacks genes encoding methanogenesis enzymes such as methyl-coenzyme M reductase, heterodisulfide reductases and hydrogenases. In situ transcriptomic analyses reveal high expression of a multi-heme c-type cytochrome, and heterologous expression of this protein in a model bacterium demonstrates that it is capable of accepting electrons. Our results suggest that Met12, within the order Methanocellales, is not a methanogen but a CO2-reducing, electron-fueled acetogen without electron bifurcation.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Deconstructing Methanosarcina acetivorans into an acetogenic archaeon.Proc Natl Acad Sci U S A. 2022 Jan 11;119(2):e2113853119. doi: 10.1073/pnas.2113853119. Proc Natl Acad Sci U S A. 2022. PMID: 34992140 Free PMC article.
-
The novel regulator HdrR controls the transcription of the heterodisulfide reductase operon hdrBCA in Methanosarcina barkeri.Appl Environ Microbiol. 2024 Jun 18;90(6):e0069124. doi: 10.1128/aem.00691-24. Epub 2024 May 29. Appl Environ Microbiol. 2024. PMID: 38809047 Free PMC article.
-
Protein complexing in a methanogen suggests electron bifurcation and electron delivery from formate to heterodisulfide reductase.Proc Natl Acad Sci U S A. 2010 Jun 15;107(24):11050-5. doi: 10.1073/pnas.1003653107. Epub 2010 Jun 1. Proc Natl Acad Sci U S A. 2010. PMID: 20534465 Free PMC article.
-
Enzymology of one-carbon metabolism in methanogenic pathways.FEMS Microbiol Rev. 1999 Jan;23(1):13-38. doi: 10.1111/j.1574-6976.1999.tb00390.x. FEMS Microbiol Rev. 1999. PMID: 10077852 Review.
-
Several ways one goal-methanogenesis from unconventional substrates.Appl Microbiol Biotechnol. 2020 Aug;104(16):6839-6854. doi: 10.1007/s00253-020-10724-7. Epub 2020 Jun 15. Appl Microbiol Biotechnol. 2020. PMID: 32542472 Free PMC article. Review.
Cited by
-
Microbial ecology of serpentinite-hosted ecosystems.ISME J. 2025 Jan 2;19(1):wraf029. doi: 10.1093/ismejo/wraf029. ISME J. 2025. PMID: 39961017 Free PMC article. Review.
-
Chemical Antiquity in Metabolism.Acc Chem Res. 2024 Aug 20;57(16):2267-2278. doi: 10.1021/acs.accounts.4c00226. Epub 2024 Jul 31. Acc Chem Res. 2024. PMID: 39083571 Free PMC article.
-
Candidate Phyla Radiation (CPR) bacteria from hyperalkaline ecosystems provide novel insight into their symbiotic lifestyle and ecological implications.Microbiome. 2025 Apr 7;13(1):94. doi: 10.1186/s40168-025-02077-y. Microbiome. 2025. PMID: 40189564 Free PMC article.
-
Evaluating Serpentinization as a Source of Phosphite to Microbial Communities in Hydrothermal Vents.Geobiology. 2025 Mar-Apr;23(2):e70016. doi: 10.1111/gbi.70016. Geobiology. 2025. PMID: 40129261 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

