Comprehensive multi-omics analysis of breast cancer reveals distinct long-term prognostic subtypes
- PMID: 38871719
- PMCID: PMC11176181
- DOI: 10.1038/s41389-024-00521-6
Comprehensive multi-omics analysis of breast cancer reveals distinct long-term prognostic subtypes
Abstract
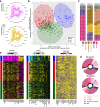
Breast cancer (BC) is a leading cause of cancer-related death worldwide. The diverse nature and heterogeneous biology of BC pose challenges for survival prediction, as patients with similar diagnoses often respond differently to treatment. Clinically relevant BC intrinsic subtypes have been established through gene expression profiling and are implemented in the clinic. While these intrinsic subtypes show a significant association with clinical outcomes, their long-term survival prediction beyond 5 years often deviates from expected clinical outcomes. This study aimed to identify naturally occurring long-term prognostic subgroups of BC based on an integrated multi-omics analysis. This study incorporates a clinical cohort of 335 untreated BC patients from the Oslo2 study with long-term follow-up (>12 years). Multi-Omics Factor Analysis (MOFA+) was employed to integrate transcriptomic, proteomic, and metabolomic data obtained from the tumor tissues. Our analysis revealed three prominent multi-omics clusters of BC patients with significantly different long-term prognoses (p = 0.005). The multi-omics clusters were validated in two independent large cohorts, METABRIC and TCGA. Importantly, a lack of prognostic association to long-term follow-up above 12 years in the previously established intrinsic subtypes was shown for these cohorts. Through a systems-biology approach, we identified varying enrichment levels of cell-cycle and immune-related pathways among the prognostic clusters. Integrated multi-omics analysis of BC revealed three distinct clusters with unique clinical and biological characteristics. Notably, these multi-omics clusters displayed robust associations with long-term survival, outperforming the established intrinsic subtypes.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources

