Prostate cancer reshapes the secreted and extracellular vesicle urinary proteomes
- PMID: 38871730
- PMCID: PMC11176296
- DOI: 10.1038/s41467-024-49424-5
Prostate cancer reshapes the secreted and extracellular vesicle urinary proteomes
Abstract
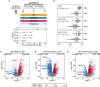
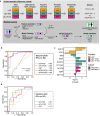
Urine is a complex biofluid that reflects both overall physiologic state and the state of the genitourinary tissues through which it passes. It contains both secreted proteins and proteins encapsulated in tissue-derived extracellular vesicles (EVs). To understand the population variability and clinical utility of urine, we quantified the secreted and EV proteomes from 190 men, including a subset with prostate cancer. We demonstrate that a simple protocol enriches prostatic proteins in urine. Secreted and EV proteins arise from different subcellular compartments. Urinary EVs are faithful surrogates of tissue proteomes, but secreted proteins in urine or cell line EVs are not. The urinary proteome is longitudinally stable over several years. It can accurately and non-invasively distinguish malignant from benign prostatic lesions and can risk-stratify prostate tumors. This resource quantifies the complexity of the urinary proteome and reveals the synergistic value of secreted and EV proteomes for translational and biomarker studies.
© 2024. The Author(s).
Conflict of interest statement
P.C.B. declares the following competing interests: P.C.B. sits on the Scientific Advisory Boards of Sage Bionetworks, Intersect Diagnostics Inc., and BioSymetrics Inc. The remaining authors declare no competing interests.
Figures





Update of
-
Prostate Cancer Reshapes the Secreted and Extracellular Vesicle Urinary Proteomes.bioRxiv [Preprint]. 2023 Jul 25:2023.07.23.550214. doi: 10.1101/2023.07.23.550214. bioRxiv. 2023. Update in: Nat Commun. 2024 Jun 13;15(1):5069. doi: 10.1038/s41467-024-49424-5. PMID: 37546794 Free PMC article. Updated. Preprint.
References
-
- Ridley, J. W. Fundamentals of the Study of Urine and Body Fluids. (Springer, Cham, Switzerland, 2018).
-
- Simerville JA, Maxted WC, Pahira JJ. Urinalysis: a comprehensive review. Am. Fam. Physician. 2005;71:1153–1162. - PubMed

