Molecular mechanisms of circular RNA translation
- PMID: 38871818
- PMCID: PMC11263353
- DOI: 10.1038/s12276-024-01220-3
Molecular mechanisms of circular RNA translation
Abstract
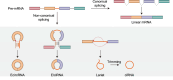
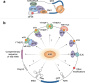
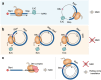
Circular RNAs (circRNAs) are covalently closed single-stranded RNAs without a 5' cap structure and a 3' poly(A) tail typically present in linear mRNAs of eukaryotic cells. CircRNAs are predominantly generated through a back-splicing process within the nucleus. CircRNAs have long been considered non-coding RNAs seemingly devoid of protein-coding potential. However, many recent studies have challenged this idea and have provided substantial evidence that a subset of circRNAs can associate with polysomes and indeed be translated. Therefore, in this review, we primarily highlight the 5' cap-independent internal initiation of translation that occurs on circular RNAs. Several molecular features of circRNAs, including the internal ribosome entry site, N6-methyladenosine modification, and the exon junction complex deposited around the back-splicing junction after back-splicing event, play pivotal roles in their efficient internal translation. We also propose a possible relationship between the translatability of circRNAs and their stability, with a focus on nonsense-mediated mRNA decay and nonstop decay, both of which are well-characterized mRNA surveillance mechanisms. An in-depth understanding of circRNA translation will reshape and expand our current knowledge of proteomics.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Circular RNAs trigger nonsense-mediated mRNA decay.Mol Cell. 2024 Dec 19;84(24):4862-4877.e7. doi: 10.1016/j.molcel.2024.11.022. Epub 2024 Dec 11. Mol Cell. 2024. PMID: 39667933
-
Translation of circular RNAs.Nucleic Acids Res. 2025 Jan 7;53(1):gkae1167. doi: 10.1093/nar/gkae1167. Nucleic Acids Res. 2025. PMID: 39660652 Free PMC article. Review.
-
How are circRNAs translated by non-canonical initiation mechanisms?Biochimie. 2019 Sep;164:45-52. doi: 10.1016/j.biochi.2019.06.015. Epub 2019 Jun 29. Biochimie. 2019. PMID: 31265859 Review.
-
Expanding uncapped translation and emerging function of circular RNA in carcinomas and noncarcinomas.Mol Cancer. 2022 Jan 7;21(1):13. doi: 10.1186/s12943-021-01484-7. Mol Cancer. 2022. PMID: 34996480 Free PMC article. Review.
-
Cellular functions and biomedical applications of circular RNAs.Acta Biochim Biophys Sin (Shanghai). 2024 Dec 24;57(1):157-168. doi: 10.3724/abbs.2024241. Acta Biochim Biophys Sin (Shanghai). 2024. PMID: 39719879 Free PMC article. Review.
Cited by
-
Deciphering the roles of non-coding RNAs in liposarcoma development: Challenges and opportunities for translational therapeutic advances.Noncoding RNA Res. 2024 Nov 15;11:73-90. doi: 10.1016/j.ncrna.2024.11.005. eCollection 2025 Apr. Noncoding RNA Res. 2024. PMID: 39736850 Free PMC article. Review.
-
RNA vaccines: The dawn of a new age for tuberculosis?Hum Vaccin Immunother. 2025 Dec;21(1):2469333. doi: 10.1080/21645515.2025.2469333. Epub 2025 Feb 27. Hum Vaccin Immunother. 2025. PMID: 40013818 Free PMC article. Review.
-
The communication role of extracellular vesicles in the osteoarthritis microenvironment.Front Immunol. 2025 Mar 17;16:1549833. doi: 10.3389/fimmu.2025.1549833. eCollection 2025. Front Immunol. 2025. PMID: 40165965 Free PMC article. Review.
-
Expanding the Potential of Circular RNA (CircRNA) Vaccines: A Promising Therapeutic Approach.Int J Mol Sci. 2025 Jan 4;26(1):379. doi: 10.3390/ijms26010379. Int J Mol Sci. 2025. PMID: 39796233 Free PMC article. Review.
-
Circular RNAs as disease modifiers of complex neurologic disorders.Front Pharmacol. 2025 May 16;16:1577496. doi: 10.3389/fphar.2025.1577496. eCollection 2025. Front Pharmacol. 2025. PMID: 40453669 Free PMC article. Review.
References
-
- Liu, C. X. & Chen, L. L. Circular RNAs: characterization, cellular roles, and applications. Cell185, 2016–2034 (2022). - PubMed
-
- Chen, L. L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol.17, 205–211 (2016). - PubMed
-
- Zhang, X. O. et al. Complementary sequence-mediated exon circularization. Cell159, 134–147 (2014). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

