Notum regulates the cusp and root patterns in mouse molar
- PMID: 38871845
- PMCID: PMC11176191
- DOI: 10.1038/s41598-024-64340-w
Notum regulates the cusp and root patterns in mouse molar
Abstract
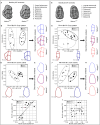
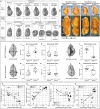
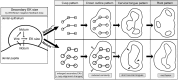
Notum is a direct target of Wnt/β-catenin signaling and plays a crucial role as a Wnt inhibitor within a negative feedback loop. In the tooth, Notum is known to be expressed in odontoblasts, and severe dentin defects and irregular tooth roots have been reported in Notum-deficient mice. However, the precise expression pattern of Notum in early tooth development, and the role of Notum in crown and root patterns remain elusive. In the present study, we identified a novel Notum expression in primary enamel knot (EK), secondary EKs, and dental papilla during tooth development. Notum-deficient mice exhibited enlarged secondary EKs, resulting in broader cusp tips, altered cusp patterns, and reduced concavity in crown outline. These alterations in crown outline led to a reduction in cervical tongue length, thereby inducing root fusion in Notum-deficient mice. Overall, these results suggest that the secondary EK size, regulated by the Wnt/Notum negative feedback loop, has a significant impact on the patterns of crown and root during tooth morphogenesis.
Keywords: Molar; Morphogenesis; Notum protein; Tooth crown; Tooth root; Wnt signaling.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Wntless regulates dentin apposition and root elongation in the mandibular molar.J Dent Res. 2015 Mar;94(3):439-45. doi: 10.1177/0022034514567198. Epub 2015 Jan 16. J Dent Res. 2015. PMID: 25595365 Free PMC article.
-
Runx2 Regulates Mouse Tooth Root Development Via Activation of WNT Inhibitor NOTUM.J Bone Miner Res. 2020 Nov;35(11):2252-2264. doi: 10.1002/jbmr.4120. Epub 2020 Aug 3. J Bone Miner Res. 2020. PMID: 32569388 Free PMC article.
-
Excessive Wnt/β-catenin signaling disturbs tooth-root formation.J Periodontal Res. 2013 Aug;48(4):405-10. doi: 10.1111/jre.12018. Epub 2012 Oct 11. J Periodontal Res. 2013. PMID: 23050778
-
Enamel knots as signaling centers linking tooth morphogenesis and odontoblast differentiation.Adv Dent Res. 2001 Aug;15:14-8. doi: 10.1177/08959374010150010401. Adv Dent Res. 2001. PMID: 12640732 Review.
-
Signaling Pathways Critical for Tooth Root Formation.J Dent Res. 2017 Oct;96(11):1221-1228. doi: 10.1177/0022034517717478. Epub 2017 Jun 30. J Dent Res. 2017. PMID: 28665752 Free PMC article. Review.
Cited by
-
Notum as a Crucial Regulator of Matrix Integrity in Dentinogenesis.J Cell Physiol. 2025 Jul;240(7):e70070. doi: 10.1002/jcp.70070. J Cell Physiol. 2025. PMID: 40685976 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

