Construction and Bioinformatics Analysis of ceRNA Regulatory Networks in Idiopathic Pulmonary Fibrosis
- PMID: 38871957
- PMCID: PMC12271262
- DOI: 10.1007/s10528-024-10853-y
Construction and Bioinformatics Analysis of ceRNA Regulatory Networks in Idiopathic Pulmonary Fibrosis
Abstract
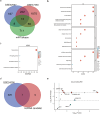
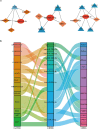
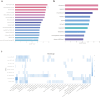
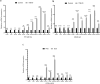
Idiopathic pulmonary fibrosis (IPF) is a chronic, progressive form of pulmonary fibrosis of unknown etiology. Despite ongoing research, there is currently no cure for this disease. Recent studies have highlighted the significance of competitive endogenous RNA (ceRNA) regulatory networks in IPF development. Therefore, this study investigated the ceRNA network associated with IPF pathogenesis. We obtained gene expression datasets (GSE32538, GSE32537, GSE47460, and GSE24206) from the Gene Expression Omnibus (GEO) database and analyzed them using bioinformatics tools to identify differentially expressed messenger RNAs (DEmRNAs), microRNAs (DEmiRNAs), and long non-coding RNAs (DElncRNA). For DEmRNAs, we conducted an enrichment analysis, constructed protein-protein interaction networks, and identified hub genes. Additionally, we predicted the target genes of differentially expressed mRNAs and their interacting long non-coding RNAs using various databases. Subsequently, we screened RNA molecules with ceRNA regulatory relations in the lncACTdb database based on the screening results. Furthermore, we performed disease and functional enrichment analyses and pathway prediction for miRNAs in the ceRNA network. We also validated the expression levels of candidate DEmRNAs through quantitative real-time reverse transcriptase polymerase chain reaction and analyzed the correlation between the expression of these candidate DEmRNAs and the percent predicted pre-bronchodilator forced vital capacity [%predicted FVC (pre-bd)]. We found that three ceRNA regulatory axes, specifically KCNQ1OT1/XIST/NEAT1-miR-20a-5p-ITGB8, XIST-miR-146b-5p/miR-31-5p- MMP16, and NEAT1-miR-31-5p-MMP16, have the potential to significantly affect IPF progression. Further examination of the underlying regulatory mechanisms within this network enhances our understanding of IPF pathogenesis and may aid in the identification of diagnostic biomarkers and therapeutic targets.
Keywords: Differential expression analysis; Pulmonary fibrosis; ceRNA; lncRNA; miRNA.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Conflict of interest: The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. Ethical Approval: The study was conducted in accordance with the Declaration of Helsinki and approved by the Guizhou Medical University Ethical Review Committee.
Figures





Similar articles
-
Identification and Characterization of a ceRNA Regulatory Network Involving LINC00482 and PRRC2B in Peripheral Blood Mononuclear Cells: Implications for COPD Pathogenesis and Diagnosis.Int J Chron Obstruct Pulmon Dis. 2024 Feb 8;19:419-430. doi: 10.2147/COPD.S437046. eCollection 2024. Int J Chron Obstruct Pulmon Dis. 2024. PMID: 38348310 Free PMC article.
-
Construction of dynamic ceRNA regulatory networks in osteogenesis during fracture healing based on transcriptomic analysis.Sci Rep. 2025 Jul 24;15(1):26946. doi: 10.1038/s41598-025-12505-6. Sci Rep. 2025. PMID: 40707643 Free PMC article.
-
Identifying pyroptosis- and inflammation-related genes in spinal cord injury based on bioinformatics analysis.Sci Rep. 2025 Jul 14;15(1):25424. doi: 10.1038/s41598-025-10541-w. Sci Rep. 2025. PMID: 40659785 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Competing endogenous RNAs in head and neck squamous cell carcinoma: a review.Brief Funct Genomics. 2024 Jul 19;23(4):335-348. doi: 10.1093/bfgp/elad049. Brief Funct Genomics. 2024. PMID: 37941447 Review.
References
-
- Anonymous. American Thoracic Society (2000) Idiopathic pulmonary fibrosis: diagnosis and treatment. International Consensus Statement. American Thoracic Society (ATS), and the European Respiratory Society (ERS). Am J Respir Crit Care Med. 161(2 Pt 1):646–664. 10.1164/ajrccm.161.2.ats3-00. - PubMed
-
- Bagnato GL, Irrera N, Pizzino G et al (2018) Dual Αvβ3 and Αvβ5 blockade attenuates fibrotic and vascular alterations in a murine model of systemic sclerosis. Clin Sci (Lond) 132(2):231–242. 10.1042/CS20171426 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

