Eomes expression identifies the early bone marrow precursor to classical NK cells
- PMID: 38871999
- PMCID: PMC11409033
- DOI: 10.1038/s41590-024-01861-6
Eomes expression identifies the early bone marrow precursor to classical NK cells
Abstract
Natural killer (NK) cells traffic through the blood and mount cytolytic and interferon-γ (IFNγ)-focused responses to intracellular pathogens and tumors. Type 1 innate lymphoid cells (ILC1s) also produce type 1 cytokines but reside in tissues and are not cytotoxic. Whether these differences reflect discrete lineages or distinct states of a common cell type is not understood. Using single-cell RNA sequencing and flow cytometry, we focused on populations of TCF7+ cells that contained precursors for NK cells and ILC1s and identified a subset of bone marrow lineage-negative NK receptor-negative cells that expressed the transcription factor Eomes, termed EomeshiNKneg cells. Transfer of EomeshiNKneg cells into Rag2-/-Il2rg-/- recipients generated functional NK cells capable of preventing metastatic disease. By contrast, transfer of PLZF+ ILC precursors generated a mixture of ILC1s, ILC2s and ILC3s that lacked cytotoxic potential. These findings identified EomeshiNKneg cells as the bone marrow precursor to classical NK cells and demonstrated that the NK and ILC1 lineages diverged early during development.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing Interests
The authors declare no competing interests.
Figures


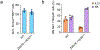









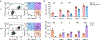
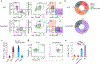


References
-
- Vivier E et al. Innate Lymphoid Cells: 10 Years On. Cell 174, 1054–1066 (2018). - PubMed
MeSH terms
Substances
Grants and funding
- R37 AI038339/AI/NIAID NIH HHS/United States
- T32 DK007074/DK/NIDDK NIH HHS/United States
- T32 AI007090/AI/NIAID NIH HHS/United States
- R01 AI038339/AI/NIAID NIH HHS/United States
- 5R01-AI144094/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- R01 AI144094/AI/NIAID NIH HHS/United States
- T32 DK007074-47/U.S. Department of Health & Human Services | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (National Institute of Diabetes & Digestive & Kidney Diseases)
- 2140001/National Science Foundation (NSF)
- P30-DK42086/UChicago | Digestive Diseases Reseach Core Center, University of Chicago (DDRCC, Uchicago)
- P30 DK042086/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

