Distinct developmental pathways generate functionally distinct populations of natural killer cells
- PMID: 38872000
- PMCID: PMC12338846
- DOI: 10.1038/s41590-024-01865-2
Distinct developmental pathways generate functionally distinct populations of natural killer cells
Erratum in
-
Author Correction: Distinct developmental pathways generate functionally distinct populations of natural killer cells.Nat Immunol. 2024 Aug;25(8):1507. doi: 10.1038/s41590-024-01917-7. Nat Immunol. 2024. PMID: 38987653 No abstract available.
Abstract
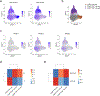
Natural killer (NK) cells function by eliminating virus-infected or tumor cells. Here we identified an NK-lineage-biased progenitor population, referred to as early NK progenitors (ENKPs), which developed into NK cells independently of common precursors for innate lymphoid cells (ILCPs). ENKP-derived NK cells (ENKP_NK cells) and ILCP-derived NK cells (ILCP_NK cells) were transcriptionally different. We devised combinations of surface markers that identified highly enriched ENKP_NK and ILCP_NK cell populations in wild-type mice. Furthermore, Ly49H+ NK cells that responded to mouse cytomegalovirus infection primarily developed from ENKPs, whereas ILCP_NK cells were better IFNγ producers after infection with Salmonella and herpes simplex virus. Human CD56dim and CD56bright NK cells were transcriptionally similar to ENKP_NK cells and ILCP_NK cells, respectively. Our findings establish the existence of two pathways of NK cell development that generate functionally distinct NK cell subsets in mice and further suggest these pathways may be conserved in humans.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Competing interests
Authors declare that they have no competing interests.
Figures

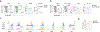



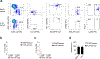

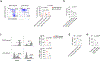

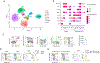

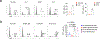

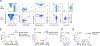

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

