Engineered plants provide a photosynthetic platform for the production of diverse human milk oligosaccharides
- PMID: 38872016
- PMCID: PMC11199141
- DOI: 10.1038/s43016-024-00996-x
Engineered plants provide a photosynthetic platform for the production of diverse human milk oligosaccharides
Abstract
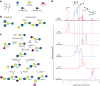
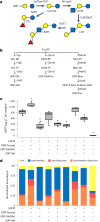
Human milk oligosaccharides (HMOs) are a diverse class of carbohydrates which support the health and development of infants. The vast health benefits of HMOs have made them a commercial target for microbial production; however, producing the approximately 200 structurally diverse HMOs at scale has proved difficult. Here we produce a diversity of HMOs by leveraging the robust carbohydrate anabolism of plants. This diversity includes high-value and complex HMOs, such as lacto-N-fucopentaose I. HMOs produced in transgenic plants provided strong bifidogenic properties, indicating their ability to serve as a prebiotic supplement with potential applications in adult and infant health. Technoeconomic analyses demonstrate that producing HMOs in plants provides a path to the large-scale production of specific HMOs at lower prices than microbial production platforms. Our work demonstrates the promise in leveraging plants for the low-cost and sustainable production of HMOs.
© 2024. The Author(s).
Conflict of interest statement
D.A.M., D.B. and C.B.L. are cofounders of Infinant Health, a company focused on probiotic-based manipulation of the infant gut microbiota. D.A.M. and C.B.L. are cofounders of One.Bio, a company advancing new bioactive glycans. None of these companies had any role in the conceptualization, design, analysis or preparation of this manuscript. The other authors declare no competing interests.
Figures



Update of
-
Plant-based production of diverse human milk oligosaccharides.bioRxiv [Preprint]. 2023 Sep 20:2023.09.18.558286. doi: 10.1101/2023.09.18.558286. bioRxiv. 2023. Update in: Nat Food. 2024 Jun;5(6):480-490. doi: 10.1038/s43016-024-00996-x. PMID: 37786679 Free PMC article. Updated. Preprint.
Similar articles
-
Plant-based production of diverse human milk oligosaccharides.bioRxiv [Preprint]. 2023 Sep 20:2023.09.18.558286. doi: 10.1101/2023.09.18.558286. bioRxiv. 2023. Update in: Nat Food. 2024 Jun;5(6):480-490. doi: 10.1038/s43016-024-00996-x. PMID: 37786679 Free PMC article. Updated. Preprint.
-
Human milk oligosaccharides and their association with late-onset neonatal sepsis in Peruvian very-low-birth-weight infants.Am J Clin Nutr. 2020 Jul 1;112(1):106-112. doi: 10.1093/ajcn/nqaa102. Am J Clin Nutr. 2020. PMID: 32401307 Free PMC article. Clinical Trial.
-
Microbial Production of Human Milk Oligosaccharides.Molecules. 2023 Feb 3;28(3):1491. doi: 10.3390/molecules28031491. Molecules. 2023. PMID: 36771155 Free PMC article. Review.
-
Diversification of a Fucosyllactose Transporter within the Genus Bifidobacterium.Appl Environ Microbiol. 2022 Jan 25;88(2):e0143721. doi: 10.1128/AEM.01437-21. Epub 2021 Nov 3. Appl Environ Microbiol. 2022. PMID: 34731055 Free PMC article.
-
In Love with Shaping You-Influential Factors on the Breast Milk Content of Human Milk Oligosaccharides and Their Decisive Roles for Neonatal Development.Nutrients. 2020 Nov 20;12(11):3568. doi: 10.3390/nu12113568. Nutrients. 2020. PMID: 33233832 Free PMC article. Review.
Cited by
-
A comprehensive framework for the production of plant-based molecules.Nat Food. 2024 Jun;5(6):461-462. doi: 10.1038/s43016-024-00995-y. Nat Food. 2024. PMID: 38872015 No abstract available.
-
EASyMap-Guided Stepwise One-Pot Multienzyme (StOPMe) Synthesis and Multiplex Assays Identify Functional Tetraose-Core-Human Milk Oligosaccharides.JACS Au. 2025 Jan 27;5(2):822-837. doi: 10.1021/jacsau.4c01094. eCollection 2025 Feb 24. JACS Au. 2025. PMID: 40017787 Free PMC article.
-
Navigating the challenges of engineering composite specialized metabolite pathways in plants.Plant J. 2025 Mar;121(6):e70100. doi: 10.1111/tpj.70100. Plant J. 2025. PMID: 40089911 Free PMC article. Review.
References
-
- 2022 Breastfeeding Report Card (CDC, 2022); https://www.cdc.gov/breastfeeding/data/reportcard.htm
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

