The radiation of New Zealand's skinks and geckos is associated with distinct viromes
- PMID: 38872095
- PMCID: PMC11170836
- DOI: 10.1186/s12862-024-02269-4
The radiation of New Zealand's skinks and geckos is associated with distinct viromes
Abstract
Background: New Zealand is home to over 120 native endemic species of skinks and geckos that radiated over the last 20-40 million years, likely driven by the exploitation of diverse habitats formed during the Miocene. The recent radiation of animal hosts may facilitate cross-species virus transmission, likely reflecting their close genetic relationships and therefore relatively low barriers for viruses to emerge in new hosts. Conversely, as animal hosts adapt to new niches, even within specific geographic locations, so too could their viruses. Consequently, animals that have niche-specialised following radiations may be expected to harbour genetically distinct viruses. Through a metatranscriptomic analysis of eight of New Zealand's native skink and gecko species, as well as the only introduced lizard species, the rainbow skink (Lampropholis delicata), we aimed to reveal the diversity of viruses in these hosts and determine whether and how the radiation of skinks and geckos in New Zealand has impacted virus diversity and evolution.
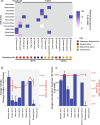
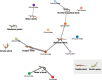
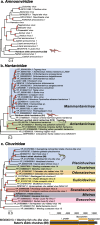
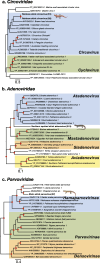
Results: We identified a total of 15 novel reptilian viruses spanning 11 different viral families, across seven of the nine species sampled. Notably, we detected no viral host-switching among the native animals analysed, even between those sampled from the same geographic location. This is compatible with the idea that host speciation has likely resulted in isolated, niche-constrained viral populations that have prevented cross-species transmission. Using a protein structural similarity-based approach, we further identified a highly divergent bunya-like virus that potentially formed a new family within the Bunyavirales.
Conclusions: This study has broadened our understanding of reptilian viruses within New Zealand and illustrates how niche adaptation may limit viral-host interactions.
Keywords: Gecko; Host radiation; Metatranscriptomics; New Zealand; Skink; Viral co-divergence; Virome.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Host and geography impact virus diversity in New Zealand's longfin and shortfin eels.Arch Virol. 2024 Mar 28;169(4):85. doi: 10.1007/s00705-024-06019-1. Arch Virol. 2024. PMID: 38546898 Free PMC article.
-
Origin, diversification, and systematics of the New Zealand skink fauna (Reptilia: Scincidae).Mol Phylogenet Evol. 2009 Aug;52(2):470-87. doi: 10.1016/j.ympev.2009.03.021. Epub 2009 Apr 2. Mol Phylogenet Evol. 2009. PMID: 19345273
-
Virome analysis of New Zealand's bats reveals cross-species viral transmission among the Coronaviridae.Virus Evol. 2024 Jan 27;10(1):veae008. doi: 10.1093/ve/veae008. eCollection 2024. Virus Evol. 2024. PMID: 38379777 Free PMC article.
-
The diverse liver viromes of Australian geckos and skinks are dominated by hepaciviruses and picornaviruses and reflect host taxonomy and habitat.Virus Evol. 2024 May 28;10(1):veae044. doi: 10.1093/ve/veae044. eCollection 2024. Virus Evol. 2024. PMID: 38854849 Free PMC article.
-
Miocene skinks and geckos reveal long-term conservatism of New Zealand's lizard fauna.Biol Lett. 2009 Dec 23;5(6):833-7. doi: 10.1098/rsbl.2009.0440. Epub 2009 Aug 5. Biol Lett. 2009. PMID: 19656857 Free PMC article.
Cited by
-
Meta-transcriptomic sequencing reveals divergent RNA viruses in geckos.Virus Res. 2025 Apr;354:199551. doi: 10.1016/j.virusres.2025.199551. Epub 2025 Feb 23. Virus Res. 2025. PMID: 39999926 Free PMC article.
-
Diversity and cross-species transmission of viruses in a remote island ecosystem: implications for wildlife conservation.Virus Evol. 2024 Dec 14;11(1):veae113. doi: 10.1093/ve/veae113. eCollection 2025. Virus Evol. 2024. PMID: 39802822 Free PMC article.
-
Avian Influenza Virus Surveillance Across New Zealand and Its Subantarctic Islands Detects H1N9 in Migratory Shorebirds, but Not 2.3.4.4b HPAI H5N1.Influenza Other Respir Viruses. 2025 Apr;19(4):e70099. doi: 10.1111/irv.70099. Influenza Other Respir Viruses. 2025. PMID: 40148670 Free PMC article.
References
-
- Uetz P, Hallermann J. The reptile database. Available from: https://reptile-database.reptarium.cz/#:~:text=Currently%20there%20are%2.... Accessed 4 Jan 2024.
-
- Hitchmough R, Barr B, Knox C, Lettink M, Monks JM, Patterson GB, Conservation status of New Zealand reptiles, , et al. Wellington: Department of Conservation. New Zeal Threat Classif Ser. 2021;2021(35):1–23.
-
- Harris J, Smith CR, van Winkel D, Brunton DH, Goulet CT, Chapple DG. Does the invasive plague skink (Lampropholis delicata) compete with native skink species in New Zealand? Austral Ecol. 2021;46(3):463–474. doi: 10.1111/aec.12984. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
