Mitochondrial DNA release via the mitochondrial permeability transition pore activates the cGAS-STING pathway, exacerbating inflammation in acute Kawasaki disease
- PMID: 38872145
- PMCID: PMC11177463
- DOI: 10.1186/s12964-024-01677-9
Mitochondrial DNA release via the mitochondrial permeability transition pore activates the cGAS-STING pathway, exacerbating inflammation in acute Kawasaki disease
Abstract
Background: Kawasaki disease (KD) is an immune vasculitis of unknown origin, characterized by transient inflammation. The activation of the cGAS-STING pathway, triggered by mitochondrial DNA (mtDNA) release, has been implicated in the onset of KD. However, its specific role in the progression of inflammation during KD's acute phase remains unclear.
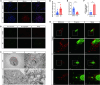
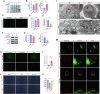
Methods: We measured mtDNA and 2'3'-cGAMP expression in KD patient serum using RT-qPCR and ELISA. A murine model of KD was induced by injecting Lactobacillus casei cell wall extract (LCWE), after which cGAS-STING pathway activation and inflammatory markers were assessed via immunohistochemistry, western blot, and RT-qPCR. Human umbilical vein endothelial cells (HUVECs) were treated with KD serum and modulators of the cGAS-STING pathway for comparative analysis. Mitochondrial function was evaluated using Mitosox staining, mPTP opening was quantified by fluorescence microscopy, and mitochondrial membrane potential (MMP) was determined with JC-1 staining.
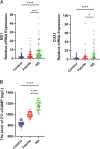
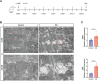
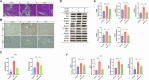
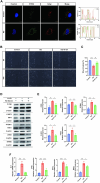
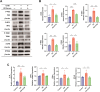
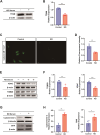
Results: KD patient serum exhibited increased mtDNA and 2'3'-cGAMP expression, with elevated levels of pathway-related proteins and inflammatory markers observed in both in vivo and in vitro models. TEM confirmed mitochondrial damage, and further studies demonstrated that inhibition of mPTP opening reduced mtDNA release, abrogated cGAS-STING pathway activation, and mitigated inflammation.
Conclusion: These findings indicate that mtDNA released through the mPTP is a critical activator of the cGAS-STING pathway, contributing significantly to KD-associated inflammation. Targeting mtDNA release or the cGAS-STING pathway may offer novel therapeutic approaches for KD management.
Keywords: Kawasaki disease; cGAS-STING pathway; mPTP; mtDNA.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Inhibition of the ATP synthase c subunit ameliorates HDM/LPS-induced inflammatory responses in asthmatic bronchial epithelial cells by blocking the mPTP-mtDNA-cGAS-STING axis.Respir Res. 2025 Jun 21;26(1):219. doi: 10.1186/s12931-025-03299-2. Respir Res. 2025. PMID: 40544259 Free PMC article.
-
Quercetin Ameliorates Learning and Memory in OVX/D-gal-Induced Alzheimer's Disease in Rats by Inhibiting Neuroinflammation via cGAS-STING Signal Pathway.Am J Chin Med. 2025;53(5):1477-1499. doi: 10.1142/S0192415X25500569. Epub 2025 Jul 15. Am J Chin Med. 2025. PMID: 40663436
-
Oxaliplatin accelerates immunogenic cell death by activating the cGAS/STING/TBK1/IRF5 pathway in gastric cancer.FEBS J. 2025 Jul;292(14):3814-3828. doi: 10.1111/febs.70102. Epub 2025 Apr 22. FEBS J. 2025. PMID: 40260556
-
Crosstalk between oxidative stress, mitochondrial dysfunction, chromosome instability, and the activation of the cGAS-STING/IFN pathway in systemic sclerosis.Ageing Res Rev. 2025 Aug;110:102812. doi: 10.1016/j.arr.2025.102812. Epub 2025 Jun 23. Ageing Res Rev. 2025. PMID: 40562314 Review.
-
Activation of the cGAS-sting Pathway Mediated by Nanocomplexes for Tumor Therapy.Curr Pharm Des. 2025;31(20):1604-1618. doi: 10.2174/0113816128339788241221160639. Curr Pharm Des. 2025. PMID: 39819537 Review.
Cited by
-
Enhancing radiotherapy-induced anti-tumor immunity via nanoparticle-mediated STING agonist synergy.Mol Cancer. 2025 Jun 11;24(1):176. doi: 10.1186/s12943-025-02366-y. Mol Cancer. 2025. PMID: 40500702 Free PMC article. Review.
-
The cGAS-STING pathway in cancer immunity: dual roles, therapeutic strategies, and clinical challenges.Essays Biochem. 2025 Mar 7;69(2):EBC20253006. doi: 10.1042/EBC20253006. Essays Biochem. 2025. PMID: 40052963 Free PMC article. Review.
-
The construction and evaluation of a prognostic risk score model for HCC based on MPT-related lncRNAs.Front Oncol. 2025 Jul 28;15:1590094. doi: 10.3389/fonc.2025.1590094. eCollection 2025. Front Oncol. 2025. PMID: 40792280 Free PMC article.
-
Mitochondrial DNA leakage: underlying mechanisms and therapeutic implications in neurological disorders.J Neuroinflammation. 2025 Feb 7;22(1):34. doi: 10.1186/s12974-025-03363-0. J Neuroinflammation. 2025. PMID: 39920753 Free PMC article. Review.
-
Redundant role of PAD2 and PAD4 in the development of cardiovascular lesions in a mouse model of Kawasaki disease vasculitis.Clin Exp Immunol. 2024 Nov 12;218(3):314-328. doi: 10.1093/cei/uxae080. Clin Exp Immunol. 2024. PMID: 39250707
References
-
- Gorelik M, Chung SA, Ardalan K, Binstadt BA, Friedman K, Hayward K, Imundo LF, Lapidus SK, Kim S, Son MB, Sule S, Tremoulet AH, Van Mater H, Yildirim-Toruner C, Langford CA, Maz M, Abril A, Guyatt G, Archer AM, Conn DL, Full KA, Grayson PC, Ibarra MF, Merkel PA, Rhee RL, Seo P, Stone JH, Sundel RP, Vitobaldi OI, Warner A, Byram K, Dua AB, Husainat N, James KE, Kalot M, Lin YC, Springer JM, Turgunbaev M, Villa-Forte A, Turner AS, Mustafa RA. 2021 American College of Rheumatology/Vasculitis Foundation Guideline for the Management of Kawasaki Disease. Arthritis Care Res (Hoboken) 2022;74(4):538–548. doi: 10.1002/acr.24838. - DOI - PubMed
-
- Nogami H, Murachi S. Phalangeal acroosteolysis associated with Down syndrome. Birth Defects Orig Artic Ser. 1975;11(2):344–346. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- SJCX23_1802/Graduate Practice Innovation Program of Jiangsu Province
- SYH-32034-0082/Jiangsu Provincial Medical Association
- 82270528/National Natural Science Foundation of China
- ZDA2020010/Jiangsu Provincial Commission of Health and Family Planning
- MS22022116/People's Livelihood Science and Technology Projetct of Nantong Science and Technology Bureau
LinkOut - more resources
Full Text Sources
Medical
Research Materials

