ALKBH5 promotes non-small cell lung cancer progression and susceptibility to anti-PD-L1 therapy by modulating interactions between tumor and macrophages
- PMID: 38872221
- PMCID: PMC11177518
- DOI: 10.1186/s13046-024-03073-0
ALKBH5 promotes non-small cell lung cancer progression and susceptibility to anti-PD-L1 therapy by modulating interactions between tumor and macrophages
Abstract
Background: Understanding the mechanisms that mediate the interaction between tumor and immune cells may provide therapeutic benefit to patients with cancer. The N6-methyladenosine (m6A) demethylase, ALKBH5 (alkB homolog 5), is overexpressed in non-small cell lung cancer. However, its role in the tumor microenvironment is unknown.
Methods: Datasets and tissue samples were used to determine the relationship between ALKBH5 expression and immunotherapy efficacy. Bioinformatic analysis, colorimetric assay to determine m6A RNA methylation, dual luciferase reporter assay, RNA/m6A-modified RNA immunoprecipitation, RNA stability assay, and RNA sequencing were used to investigate the regulatory mechanism of ALKBH5 in non-small cell lung cancer. In vitro and in vivo assays were performed to determine the contribution of ALKBH5 to the development of non-small cell lung cancer.
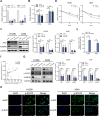
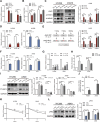
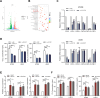
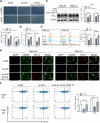
Results: ALKBH5 was upregulated in primary non-small cell lung cancer tissues. ALKBH5 was positively correlated with programmed death-ligand 1 expression and macrophage infiltration and was associated with immunotherapy response. JAK2 was identified as a target of ALKBH5-mediated m6A modification, which activates the JAK2/p-STAT3 pathway to promote non-small cell lung cancer progression. ALKBH5 was found to recruit programmed death-ligand 1-positive tumor-associated macrophages and promote M2 macrophage polarization by inducing the secretion of CCL2 and CXCL10. ALKBH5 and tumor-associated macrophage-secreted IL-6 showed a synergistic effect to activate the JAK2/p-STAT3 pathway in cancer cells.
Conclusions: ALKBH5 promotes non-small cell lung cancer progression by regulating cancer and tumor-associated macrophage behavior through the JAK2/p-STAT3 pathway and the expression of CCL2 and CXCL10, respectively. These findings suggest that targeting ALKBH5 is a promising strategy of enhancing the anti-tumor immune response in patients with NSCLC and that identifying ALKBH5 status could facilitate prediction of clinical response to anti-PD-L1 immunotherapy.
Keywords: ALKBH5; JAK2/p-STAT3 pathway; N6-methyladenosine demethylase; Non-small cell lung cancer; Tumor microenvironment; Tumor-associated macrophage.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
ALKBH5-Mediated m6A Modification of XBP1 Facilitates NSCLC Progression Through the IL-6-JAK-STAT3 Pathway.Mol Carcinog. 2025 Jan;64(1):57-71. doi: 10.1002/mc.23826. Epub 2024 Oct 10. Mol Carcinog. 2025. PMID: 39387829
-
High expression of AlkB homolog 5 suppresses the progression of non-small cell lung cancer by facilitating ferroptosis through m6A demethylation of SLC7A11.Environ Toxicol. 2024 Jul;39(7):4035-4046. doi: 10.1002/tox.24272. Epub 2024 Apr 20. Environ Toxicol. 2024. PMID: 38642004
-
M6A Demethylase ALKBH5 Regulates PD-L1 Expression and Tumor Immunoenvironment in Intrahepatic Cholangiocarcinoma.Cancer Res. 2021 Sep 15;81(18):4778-4793. doi: 10.1158/0008-5472.CAN-21-0468. Epub 2021 Jul 23. Cancer Res. 2021. PMID: 34301762
-
Unraveling molecular and clinical aspects of ALKBH5 as dual role in colorectal cancer.J Pharm Pharmacol. 2024 Nov 4;76(11):1393-1403. doi: 10.1093/jpp/rgae108. J Pharm Pharmacol. 2024. PMID: 39321327 Review.
-
Epidermal Growth Factor Receptor (EGFR) Pathway, Yes-Associated Protein (YAP) and the Regulation of Programmed Death-Ligand 1 (PD-L1) in Non-Small Cell Lung Cancer (NSCLC).Int J Mol Sci. 2019 Aug 5;20(15):3821. doi: 10.3390/ijms20153821. Int J Mol Sci. 2019. PMID: 31387256 Free PMC article. Review.
Cited by
-
Analysis of Genotype and Expression of FTO and ALKBH5 in a MENA-Region Renal Cell Carcinoma Cohort.Cancers (Basel). 2025 Apr 22;17(9):1395. doi: 10.3390/cancers17091395. Cancers (Basel). 2025. PMID: 40361322 Free PMC article.
-
NGR-modified nanovesicles target ALKBH5 to inhibit ovarian cancer growth and metastasis.Theranostics. 2025 Jun 9;15(14):6702-6718. doi: 10.7150/thno.107766. eCollection 2025. Theranostics. 2025. PMID: 40585991 Free PMC article.
-
RNA modifications in cancer immune therapy: regulators of immune cells and immune checkpoints.Front Immunol. 2024 Sep 20;15:1463847. doi: 10.3389/fimmu.2024.1463847. eCollection 2024. Front Immunol. 2024. PMID: 39372415 Free PMC article. Review.
-
Mechanistic insights into the role of RNA demethylase ALKBH5 in malignant tumor therapy.J Transl Med. 2025 Aug 13;23(1):905. doi: 10.1186/s12967-025-06938-w. J Transl Med. 2025. PMID: 40804394 Free PMC article. Review.
-
Tumor cell-intrinsic BIN1 deficiency promotes the immunosuppression and impedes ferroptosis of non-small cell lung cancer via G3BP1-mediated degradation of STAT1.J Exp Clin Cancer Res. 2025 May 9;44(1):141. doi: 10.1186/s13046-025-03404-9. J Exp Clin Cancer Res. 2025. PMID: 40346580 Free PMC article.
References
MeSH terms
Substances
Grants and funding
- 82370096/National Natural Science Foundation of China
- 82172728/National Natural Science Foundation of China
- BE2019719/Jiangsu Provincial Key Research and Development Program
- 2023JCYJZD080/Science and Technology Innovation Research Project of Jinling Hospital
- 22LCYY-LH3/Science and Technology Innovation Research Project of Jinling Hospital
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

