Metagenomic insights into isolable bacterial communities and antimicrobial resistance in airborne dust from pig farms
- PMID: 38872793
- PMCID: PMC11169874
- DOI: 10.3389/fvets.2024.1362011
Metagenomic insights into isolable bacterial communities and antimicrobial resistance in airborne dust from pig farms
Abstract
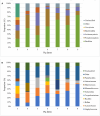
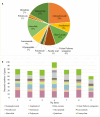
This study aims to investigate bacterial communities and antimicrobial resistance (AMR) in airborne dust from pig farms. Airborne dust, pig feces and feed were collected from nine pig farms in Thailand. Airborne dust samples were collected from upwind and downwind (25 meters from pig house), and inside (in the middle of the pig house) of the selected pig house. Pig feces and feed samples were individually collected from the pen floor and feed trough from the same pig house where airborne dust was collected. A direct total bacteria count on each sampling plate was conducted and averaged. The ESKAPE pathogens together with Escherichia coli, Salmonella, and Streptococcus were examined. A total of 163 bacterial isolates were collected and tested for MICs. Pooled bacteria from the inside airborne dust samples were analyzed using Metagenomic Sequencing. The highest bacterial concentration (1.9-11.2 × 103 CFU/m3) was found inside pig houses. Staphylococcus (n = 37) and Enterococcus (n = 36) were most frequent bacterial species. Salmonella (n = 3) were exclusively isolated from feed and feces. Target bacteria showed a variety of resistance phenotypes, and the same bacterial species with the same resistance phenotype were found in airborne dust, feed and fecal from each farm. Metagenomic Sequencing analysis revealed 1,652 bacterial species across all pig farms, of which the predominant bacterial phylum was Bacillota. One hundred fifty-nine AMR genes of 12 different antibiotic classes were identified, with aminoglycoside resistance genes (24%) being the most prevalent. A total of 251 different plasmids were discovered, and the same plasmid was detected in multiple farms. In conclusion, the phenotypic and metagenomic results demonstrated that airborne dust from pig farms contained a diverse array of bacterial species and genes encoding resistance to a range of clinically important antimicrobial agents, indicating the significant role in the spread of AMR bacterial pathogens with potential hazards to human health. Policy measurements to address AMR in airborne dust from livestock farms are mandatory.
Keywords: airborne dust; antimicrobial resistance; bacterial communities; metagenomic approach; pig farm.
Copyright © 2024 Hein, Prathan, Srisanga, Muenhor, Wongsurawat, Jenjaroenpun, Tummaruk and Chuanchuen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Farm dust resistomes and bacterial microbiomes in European poultry and pig farms.Environ Int. 2020 Oct;143:105971. doi: 10.1016/j.envint.2020.105971. Epub 2020 Jul 29. Environ Int. 2020. PMID: 32738764
-
Spread of airborne antibiotic resistance from animal farms to the environment: Dispersal pattern and exposure risk.Environ Int. 2022 Jan;158:106927. doi: 10.1016/j.envint.2021.106927. Epub 2021 Oct 18. Environ Int. 2022. PMID: 34673316
-
Determinants for antimicrobial resistance genes in farm dust on 333 poultry and pig farms in nine European countries.Environ Res. 2022 May 15;208:112715. doi: 10.1016/j.envres.2022.112715. Epub 2022 Jan 13. Environ Res. 2022. PMID: 35033551
-
Airborne bacterial community and antibiotic resistome in the swine farming environment: Metagenomic insights into livestock relevance, pathogen hosts and public risks.Environ Int. 2023 Feb;172:107751. doi: 10.1016/j.envint.2023.107751. Epub 2023 Jan 13. Environ Int. 2023. PMID: 36680804
-
Patterns of antibiotic use in global pig production: A systematic review.Vet Anim Sci. 2019 Apr 6;7:100058. doi: 10.1016/j.vas.2019.100058. eCollection 2019 Jun. Vet Anim Sci. 2019. PMID: 32734079 Free PMC article. Review.
References
-
- UNEP . Frontiers 2017 Emerging Issues of Environmental Concern. United Nations Environment Programme Nairobi; (2017).
LinkOut - more resources
Full Text Sources

