Update on functional analysis of long non-coding RNAs in common crops
- PMID: 38872885
- PMCID: PMC11169716
- DOI: 10.3389/fpls.2024.1389154
Update on functional analysis of long non-coding RNAs in common crops
Abstract
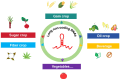
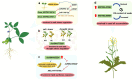
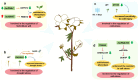
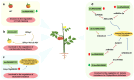
With the rapid advances in next-generation sequencing technology, numerous non-protein-coding transcripts have been identified, including long noncoding RNAs (lncRNAs), which are functional RNAs comprising more than 200 nucleotides. Although lncRNA-mediated regulatory processes have been extensively investigated in animals, there has been considerably less research on plant lncRNAs. Nevertheless, multiple studies on major crops showed lncRNAs are involved in crucial processes, including growth and development, reproduction, and stress responses. This review summarizes the progress in the research on lncRNA roles in several major crops, presents key strategies for exploring lncRNAs in crops, and discusses current challenges and future prospects. The insights provided in this review will enhance our comprehension of lncRNA functions in crops, with potential implications for improving crop genetics and breeding.
Keywords: biological function; crops; lncRNAs; molecular mechanism; plant development.
Copyright © 2024 Zhang, Pi, Wang, Li, Wang, Liu, Cui, Liu, Yao and Zhao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Abdel-Salam E. M., Qahtan A. A., Gaafar A. Z., Zein El-Abedein A. I., Alshameri A. M., Alhamdan A. M. (2021). Tissue-specific analysis of Coffea arabica L. transcriptome revealed potential regulatory roles of lncRNAs. Saudi J. Biol. Sci. 28, 6023–6029. doi: 10.1016/j.sjbs.2021.07.006 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources

