Biocontrol potential of endophytic Bacillus subtilis A9 against rot disease of Morchella esculenta
- PMID: 38873148
- PMCID: PMC11169702
- DOI: 10.3389/fmicb.2024.1388669
Biocontrol potential of endophytic Bacillus subtilis A9 against rot disease of Morchella esculenta
Abstract
Introduction: Morchella esculenta is a popular edible fungus with high economic and nutritional value. However, the rot disease caused by Lecanicillium aphanocladii, pose a serious threat to the quality and yield of M. esculenta. Biological control is one of the effective ways to control fungal diseases.
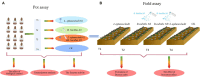
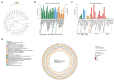
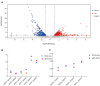
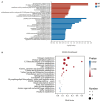
Methods and results: In this study, an effective endophytic B. subtilis A9 for the control of M. esculenta rot disease was screened, and its biocontrol mechanism was studied by transcriptome analysis. In total, 122 strains of endophytic bacteria from M. esculenta, of which the antagonistic effect of Bacillus subtilis A9 on L. aphanocladii G1 reached 72.2% in vitro tests. Biological characteristics and genomic features of B. subtilis A9 were analyzed, and key antibiotic gene clusters were detected. Scanning electron microscope (SEM) observation showed that B. subtilis A9 affected the mycelium and spores of L. aphanocladii G1. In field experiments, the biological control effect of B. subtilis A9 reached to 62.5%. Furthermore, the transcritome profiling provides evidence of B. subtilis A9 bicontrol at the molecular level. A total of 1,246 differentially expressed genes (DEGs) were identified between the treatment and control group. Gene Ontology (GO) enrichment analysis showed that a large number of DEGs were related to antioxidant activity related. Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis showed that the main pathways were Nitrogen metabolism, Pentose Phosphate Pathway (PPP) and Mitogen-Activated Protein Kinases (MAPK) signal pathway. Among them, some important genes such as carbonic anhydrase CA (H6S33_007248), catalase CAT (H6S33_001409), tRNA dihydrouridine synthase DusB (H6S33_001297) and NAD(P)-binding protein NAD(P) BP (H6S33_000823) were found. Furthermore, B. subtilis A9 considerably enhanced the M. esculenta activity of Polyphenol oxidase (POD), Superoxide dismutase (SOD), Phenylal anineammonia lyase (PAL) and Catalase (CAT).
Conclusion: This study presents the innovative utilization of B. subtilis A9, for effectively controlling M. esculenta rot disease. This will lay a foundation for biological control in Morchella, which may lead to the improvement of new biocontrol agents for production.
Keywords: Lecanicillium aphanocladii; Morchella esculenta; biological control; fungal disease; transcriptome analysis.
Copyright © 2024 Chen, Zhang, Chao, Song, Wu, Sun, Chen and Lv.
Conflict of interest statement
BL was employed by Shanghai Co-Elite Agricultural Sci-Tech (Group) Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Home treatment for mental health problems: a systematic review.Health Technol Assess. 2001;5(15):1-139. doi: 10.3310/hta5150. Health Technol Assess. 2001. PMID: 11532236
-
Preliminary study on Cyclocodon lancifolius leaf blight and screening of Bacillus subtilis as a biocontrol agent.Front Microbiol. 2024 Oct 15;15:1459868. doi: 10.3389/fmicb.2024.1459868. eCollection 2024. Front Microbiol. 2024. PMID: 39473852 Free PMC article.
-
Plant Growth Promotion and Biocontrol of Leaf Blight Caused by Nigrospora sphaerica on Passion Fruit by Endophytic Bacillus subtilis Strain GUCC4.J Fungi (Basel). 2023 Jan 18;9(2):132. doi: 10.3390/jof9020132. J Fungi (Basel). 2023. PMID: 36836247 Free PMC article.
-
Understanding mechanisms of Polygonatum sibiricum-derived exosome-like nanoparticles against breast cancer through an integrated metabolomics and network pharmacology analysis.Front Chem. 2025 Jun 6;13:1559758. doi: 10.3389/fchem.2025.1559758. eCollection 2025. Front Chem. 2025. PMID: 40547857 Free PMC article.
Cited by
-
Enhancing cellulase biosynthesis of Bacillus subtilis Z2 by regulating intracellular NADH level.iScience. 2025 Apr 3;28(5):112341. doi: 10.1016/j.isci.2025.112341. eCollection 2025 May 16. iScience. 2025. PMID: 40276757 Free PMC article.
-
Streptomyces-Based Bioformulation to Control Wilt of Morchella sextelata Caused by Pestalotiopsis trachicarpicola.J Fungi (Basel). 2025 Jun 13;11(6):452. doi: 10.3390/jof11060452. J Fungi (Basel). 2025. PMID: 40558963 Free PMC article.
References
-
- Abdelkhalek A., Behiry S. I., Al-Askar A. A. (2020). Bacillus velezensis PEA1 inhibits Fusarium oxysporum growth and induces systemic resistance to cucumber mosaic virus. Agronomy 10:1312. doi: 10.3390/agronomy10091312 - DOI
-
- Ahmed W., Yang J., Tan Y., Munir S., Liu Q., Zhang J., et al. (2022). Ralstonia solanacearum, a deadly pathogen: revisiting the bacterial wilt biocontrol practices in tobacco and other Solanaceae. Rhizosphere:100479: 21. doi: 10.1016/j.rhisph.2022.100479 - DOI
-
- Algburi A., Zehm S., Netrebov V., Bren A. B., Chistyakov V., Chikindas M. L. (2016). Subtilosin prevents biofilm formation by inhibiting bacterial quorum sensing. Probiot. Antimicrob. Proteins 9, 81–90, - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

