Blockage of TMEM189 induces G2/M arrest and inhibits the growth of breast tumors
- PMID: 38873225
- PMCID: PMC11170349
- DOI: 10.1016/j.bbrep.2024.101744
Blockage of TMEM189 induces G2/M arrest and inhibits the growth of breast tumors
Abstract
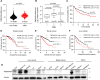
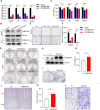
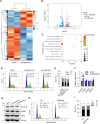
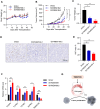
Cancer is the major cause of premature death in humans worldwide, demanding more efficient therapeutics. Aberrant cell proliferation resulting from the loss of cell cycle regulation is the major hallmark of cancer, so targeting cell cycle is a promising strategy to combat cancer. However, the molecular mechanism underlying the dysregulation of cell cycle of cancer cells remains poorly understood. TMEM189, a newly identified protein, plays roles in the biosynthesis of ethanolamine plasmalogen and the regulation of autophagy. Here, we demonstrated that the expression level of TMEM189 was negatively correlated with the survival rate of the cancer patients. TMEM189 deficiency significantly suppresses the cancer cell proliferation and migration, and causes cell cycle G2/M arrest both in vitro and in vivo. Furthermore, TMEM189 depletion suppressed the growth of breast tumors in vivo. Taken together, our work indicated that TMEM189 promotes cancer progression by regulating cell cycle G2/M transition, suggesting that it is a promising target in cancer therapy.
© 2024 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
Similar articles
-
TMEM189 promotes breast cancer through inhibition of autophagy-regulated ferroptosis.Biochem Biophys Res Commun. 2022 Sep 24;622:37-44. doi: 10.1016/j.bbrc.2022.06.024. Epub 2022 Jun 12. Biochem Biophys Res Commun. 2022. PMID: 35843092
-
TMEM189 as a target gene of MiR-499a-5p regulates breast cancer progression through the ferroptosis pathway.J Clin Biochem Nutr. 2023 Sep;73(2):154-160. doi: 10.3164/jcbn.22-130. Epub 2023 Sep 1. J Clin Biochem Nutr. 2023. PMID: 37700851 Free PMC article.
-
TMEM189 negatively regulates the stability of ULK1 protein and cell autophagy.Cell Death Dis. 2022 Apr 7;13(4):316. doi: 10.1038/s41419-022-04722-y. Cell Death Dis. 2022. PMID: 35393404 Free PMC article.
-
The Severe Fever with Thrombocytopenia Syndrome Virus NSs Protein Interacts with CDK1 To Induce G2 Cell Cycle Arrest and Positively Regulate Viral Replication.J Virol. 2020 Feb 28;94(6):e01575-19. doi: 10.1128/JVI.01575-19. Print 2020 Feb 28. J Virol. 2020. PMID: 31852787 Free PMC article.
-
Plasmalogens and Photooxidative Stress Signaling in Myxobacteria, and How it Unmasked CarF/TMEM189 as the Δ1'-Desaturase PEDS1 for Human Plasmalogen Biosynthesis.Front Cell Dev Biol. 2022 May 11;10:884689. doi: 10.3389/fcell.2022.884689. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35646900 Free PMC article. Review.
References
-
- Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12:31–46. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

