Integrated mRNA-miRNA transcriptome profiling of blood immune responses potentially related to pulmonary fibrosis in forest musk deer
- PMID: 38873601
- PMCID: PMC11169664
- DOI: 10.3389/fimmu.2024.1404108
Integrated mRNA-miRNA transcriptome profiling of blood immune responses potentially related to pulmonary fibrosis in forest musk deer
Abstract
Background: Forest musk deer (FMD, Moschus Berezovskii) is a critically endangered species world-widely, the death of which can be caused by pulmonary disease in the farm. Pulmonary fibrosis (PF) was a huge threat to the health and survival of captive FMD. MicroRNAs (miRNAs) and messenger RNAs (mRNAs) have been involved in the regulation of immune genes and disease development. However, the regulatory profiles of mRNAs and miRNAs involved in immune regulation of FMD are unclear.
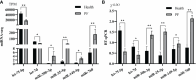
Methods: In this study, mRNA-seq and miRNA-seq in blood were performed to constructed coexpression regulatory networks between PF and healthy groups of FMD. The hub immune- and apoptosis-related genes in the PF blood of FMD were explored through Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis. Further, protein-protein interaction (PPI) network of immune-associated and apoptosis-associated key signaling pathways were constructed based on mRNA-miRNA in the PF blood of the FMD. Immune hub DEGs and immune hub DEmiRNAs were selected for experimental verification using RT-qPCR.
Results: A total of 2744 differentially expressed genes (DEGs) and 356 differentially expressed miRNAs (DEmiRNAs) were identified in the PF blood group compared to the healthy blood group. Among them, 42 DEmiRNAs were negatively correlated with 20 immune DEGs from a total of 57 correlations. The DEGs were significantly associated with pathways related to CD molecules, immune disease, immune system, cytokine receptors, T cell receptor signaling pathway, Th1 and Th2 cell differentiation, cytokine-cytokine receptor interaction, intestinal immune network for IgA production, and NOD-like receptor signaling pathway. There were 240 immune-related DEGs, in which 186 immune-related DEGs were up-regulated and 54 immune-related DEGs were down-regulated. In the protein-protein interaction (PPI) analysis of immune-related signaling pathway, TYK2, TLR2, TLR4, IL18, CSF1, CXCL13, LCK, ITGB2, PIK3CB, HCK, CD40, CD86, CCL3, CCR7, IL2RA, TLR3, and IL4R were identified as the hub immune genes. The mRNA-miRNA coregulation analysis showed that let-7d, miR-324-3p, miR-760, miR-185, miR-149, miR-149-5p, and miR-1842-5p are key miRNAs that target DEGs involved in immune disease, immune system and immunoregulation.
Conclusion: The development and occurrence of PF were significantly influenced by the immune-related and apoptosis-related genes present in PF blood. mRNAs and miRNAs associated with the development and occurrence of PF in the FMD.
Keywords: RT-qPCR; blood transcriptome; forest musk deer; immune response; miRNA-mRNA network; signal pathway analysis.
Copyright © 2024 Qi, Hu, Gu, Zhang, Jiang, Li, Qi, Xiao and Jie.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
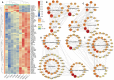


References
-
- Yang Q, Meng X, Xia L, Feng Z. Conservation status and causes of decline of musk deer (Moschus spp.) in China. Biol Conserv. (2003) 109:333–42. doi: 10.1016/S0006-3207(02)00159-3 - DOI
-
- Shrestha M. Animal welfare in the musk deer. Appl Anim Behav Sci. (1998) 59:245–50. doi: 10.1016/S0168-1591(98)00139-7 - DOI
-
- Meng X, Gong B, Ma G, Xiang L. Quantified analyses of musk deer farming in China: a tool for sustainable musk production and ex situ conservation. Asian Australas J Anim Sci. (2011) 24:1473–82. doi: 10.5713/ajas.2011.11111 - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

