The autophagy protein RUBCNL/PACER represses RIPK1 kinase-dependent apoptosis and necroptosis
- PMID: 38873940
- PMCID: PMC11572279
- DOI: 10.1080/15548627.2024.2367923
The autophagy protein RUBCNL/PACER represses RIPK1 kinase-dependent apoptosis and necroptosis
Abstract
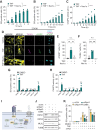
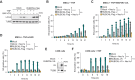
Mesenchymal stem cells (MSCs) are used in cell therapy; nonetheless, their application is limited by their poor survival after transplantation in a proinflammatory microenvironment. Macroautophagy/autophagy activation in MSCs constitutes a stress adaptation pathway, promoting cellular homeostasis. Our proteomics data indicate that RUBCNL/PACER (RUN and cysteine rich domain containing beclin 1 interacting protein like), a positive regulator of autophagy, is also involved in cell death. Hence, we screened MSC survival upon various cell death stimuli under loss or gain of function of RUBCNL. MSCs were protected from TNF (tumor necrosis factor)-induced regulated cell death when RUBCNL was expressed. TNF promotes inflammation by inducing RIPK1 kinase-dependent apoptosis or necroptosis. We determine that MSCs succumb to RIPK1 kinase-dependent apoptosis upon TNF sensing and necroptosis when caspases are inactivated. We show that RUBCNL is a negative regulator of both RIPK1-dependent apoptosis and necroptosis. Furthermore, RUBCNL mutants that lose the ability to regulate autophagy, retain their function in negatively regulating cell death. We also found that RUBCNL forms a complex with RIPK1, which disassembles in response to TNF. In line with this finding, RUBCNL expression limits assembly of RIPK1-TNFRSF1A/TNFR1 complex I, suggesting that complex formation between RUBCNL and RIPK1 represses TNF signaling. These results provide new insights into the crosstalk between the RIPK1-mediated cell death and autophagy machineries and suggest that RUBCNL, due to its functional duality in autophagy and apoptosis/necroptosis, could be targeted to improve the therapeutic efficacy of MSCs. Abbreviations: BAF: bafilomycin A1; CASP3: caspase 3; Caspases: cysteine-aspartic proteases; cCASP3: cleaved CASP3; CQ: chloroquine; CHX: cycloheximide; cPARP: cleaved poly (ADP-ribose) polymerase; DEPs: differential expressed proteins; ETO: etoposide; MEF: mouse embryonic fibroblast; MLKL: mixed lineage kinase domain-like; MSC: mesenchymal stem cell; MTORC1: mechanistic target of rapamycin kinase complex 1; Nec1s: necrostatin 1s; NFKB/NF-kB: nuclear factor of kappa light polypeptide gene enhancer in B cells; PLA: proximity ligation assay; RCD: regulated cell death; RIPK1: receptor (TNFRSF)-interacting serine-threonine kinase 1; RIPK3: receptor-interacting serine-threonine kinase 3; RUBCNL/PACER: RUN and cysteine rich domain containing beclin 1 interacting protein like; siCtrl: small interfering RNA nonsense; siRNA: small interfering RNA; TdT: terminal deoxynucleotidyl transferase; Tm: tunicamycin; TNF: tumor necrosis factor; TNFRSF1A/TNFR1: tumor necrosis factor receptor superfamily, member 1a.
Keywords: Cell death; KIAA0226L; TNF; TNFR1; mesenchymal stem cells; necrostatin 1s.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures




References
-
- Liu X, Chen H, Chen H, et al. Angiopoietin-1 preconditioning enhances survival and functional recovery of mesenchymal stem cell transplantation. J Zhejiang Univ Sci B [Internet]. 2012. [cited 2018 May 21];13(8):616–623. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22843181 - PMC - PubMed
-
- Li L, Chen X, Wang WE, et al. How to improve the survival of transplanted mesenchymal stem cell in ischemic heart? STEM Cells Int [Internet]. 2016. [cited 2018 May 21];2016:9682757. Available from: http://www.hindawi.com/journals/sci/2016/9682757/ - PMC - PubMed
-
- Prockop DJ. Repair of tissues by adult stem/progenitor cells (MSCs): controversies, myths, and changing paradigms. Mol Ther [Internet]. 2009. [cited 2022 Jan 11];17(6):939–946. Available from: https://pubmed.ncbi.nlm.nih.gov/19337235/ - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
