Overcoming the High Error Rate of Composite DNA Letters-Based Digital Storage through Soft-Decision Decoding
- PMID: 38874370
- PMCID: PMC11321706
- DOI: 10.1002/advs.202402951
Overcoming the High Error Rate of Composite DNA Letters-Based Digital Storage through Soft-Decision Decoding
Abstract
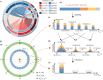
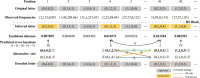
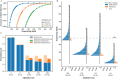
Composite DNA letters, by merging all four DNA nucleotides in specified ratios, offer a pathway to substantially increase the logical density of DNA digital storage (DDS) systems. However, these letters are susceptible to nucleotide errors and sampling bias, leading to a high letter error rate, which complicates precise data retrieval and augments reading expenses. To address this, Derrick-cp is introduced as an innovative soft-decision decoding algorithm tailored for DDS utilizing composite letters. Derrick-cp capitalizes on the distinctive error sensitivities among letters to accurately predict and rectify letter errors, thus enhancing the error-correcting performance of Reed-Solomon codes beyond traditional hard-decision decoding limits. Through comparative analyses in the existing dataset and simulated experiments, Derrick-cp's superiority is validated, notably halving the sequencing depth requirement and slashing costs by up to 22% against conventional hard-decision strategies. This advancement signals Derrick-cp's significant role in elevating both the precision and cost-efficiency of composite letter-based DDS.
Keywords: DNA digital storage (DDS); composite DNA letter; error‐correcting code (ECC); soft‐decision decoding.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Improving error-correcting capability in DNA digital storage via soft-decision decoding.Natl Sci Rev. 2023 Sep 2;11(2):nwad229. doi: 10.1093/nsr/nwad229. eCollection 2024 Feb. Natl Sci Rev. 2023. PMID: 38213525 Free PMC article.
-
Iterative Soft Decoding Algorithm for DNA Storage Using Quality Score and Redecoding.IEEE Trans Nanobioscience. 2024 Jan;23(1):81-90. doi: 10.1109/TNB.2023.3284406. Epub 2024 Jan 3. IEEE Trans Nanobioscience. 2024. PMID: 37294652
-
Data storage in DNA with fewer synthesis cycles using composite DNA letters.Nat Biotechnol. 2019 Oct;37(10):1229-1236. doi: 10.1038/s41587-019-0240-x. Epub 2019 Sep 9. Nat Biotechnol. 2019. PMID: 31501560
-
Efficient data reconstruction: The bottleneck of large-scale application of DNA storage.Cell Rep. 2024 Apr 23;43(4):113699. doi: 10.1016/j.celrep.2024.113699. Epub 2024 Mar 21. Cell Rep. 2024. PMID: 38517891 Review.
-
Recent progress in DNA data storage based on high-throughput DNA synthesis.Biomed Eng Lett. 2024 May 3;14(5):993-1009. doi: 10.1007/s13534-024-00386-z. eCollection 2024 Sep. Biomed Eng Lett. 2024. PMID: 39220021 Free PMC article. Review.
Cited by
-
Pragmatic soft-decision data readout of encoded large DNA.Brief Bioinform. 2025 Mar 4;26(2):bbaf102. doi: 10.1093/bib/bbaf102. Brief Bioinform. 2025. PMID: 40091194 Free PMC article.
References
-
- a) Dong Y., Sun F., Ping Z., Ouyang Q., Qian L., Nat. Sci. Rev. 2020, 7, 1092; - PMC - PubMed
- b) Meiser L. C., Antkowiak P. L., Koch J., Chen W. D., Kohll A. X., Stark W. J., Heckel R., Grass R. N., Nat. Protoc. 2020, 15, 86; - PubMed
- c) Ping Z., Ma D., Huang X., Chen S., Liu L., Guo F., Zhu S. J., Shen Y., GigaScience 2019, 8, giz075; - PMC - PubMed
- d) Zhirnov V., Zadegan R. M., Sandhu G. S., Church G. M., Hughes W. L., Nat. Mater. 2016, 15, 366. - PMC - PubMed
-
- a) Nguyen T. T., Cai K., Immink K. A. S., Kiah H. M., in IEEE International Symposium on Information Theory (ISIT) 2020, 694;
- b) Wang Y., Noor‐A‐Rahim M., Gunawan E., Guan Y. L., Poh C. L., IEEE Communications Letters 2019, 23, 963;
- c) Ceze L., Nivala J., Strauss K., Nat. Rev. Genet. 2019, 20, 456. - PubMed
-
- Anavy L., Vaknin I., Atar O., Amit R., Yakhini Z., Nat. Biotechnol. 2019, 37, 1229. - PubMed
-
- a) Chen Y.‐J., Takahashi C. N., Organick L., Bee C., Ang S. D., Weiss P., Peck B., Seelig G., Ceze L., Strauss K., Nat. Commun. 2020, 11, 3264; - PMC - PubMed
- b) Organick L., Ang S. D., Chen Y.‐J., Lopez R., Yekhanin S., Makarychev K., Racz M. Z., Kamath G., Gopalan P., Nguyen B., Takahashi C. N., Newman S., Parker H.‐Y., Rashtchian C., Stewart K., Gupta G., Carlson R., Mulligan J., Carmean D., Seelig G., Ceze L., Strauss K., Nat. Biotechnol. 2018, 36, 242. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
