Lefamulin Overcomes Acquired Drug Resistance via Regulating Mitochondrial Homeostasis by Targeting ILF3 in Hepatocellular Carcinoma
- PMID: 38874478
- PMCID: PMC11321631
- DOI: 10.1002/advs.202401789
Lefamulin Overcomes Acquired Drug Resistance via Regulating Mitochondrial Homeostasis by Targeting ILF3 in Hepatocellular Carcinoma
Abstract
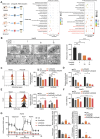
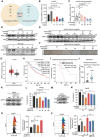
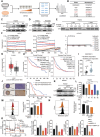
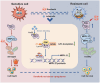
Acquired resistance represents a critical clinical challenge to molecular targeted therapies such as tyrosine kinase inhibitors (TKIs) treatment in hepatocellular carcinoma (HCC). Therefore, it is urgent to explore new mechanisms and therapeutics that can overcome or delay resistance. Here, a US Food and Drug Administration (FDA)-approved pleuromutilin antibiotic is identified that overcomes sorafenib resistance in HCC cell lines, cell line-derived xenograft (CDX) and hydrodynamic injection mouse models. It is demonstrated that lefamulin targets interleukin enhancer-binding factor 3 (ILF3) to increase the sorafenib susceptibility of HCC via impairing mitochondrial function. Mechanistically, lefamulin directly binds to the Alanine-99 site of ILF3 protein and interferes with acetyltransferase general control non-depressible 5 (GCN5) and CREB binding protein (CBP) mediated acetylation of Lysine-100 site, which disrupts the ILF3-mediated transcription of mitochondrial ribosomal protein L12 (MRPL12) and subsequent mitochondrial biogenesis. Clinical data further confirm that high ILF3 or MRPL12 expression is associated with poor survival and targeted therapy efficacy in HCC. Conclusively, this findings suggest that ILF3 is a potential therapeutic target for overcoming resistance to TKIs, and lefamulin may be a novel combination therapy strategy for HCC treatment with sorafenib and regorafenib.
Keywords: HCC; ILF3; MRPL12; acetylation; lefamulin; targeted therapy resistance.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Siegel R. L., Miller K. D., Fuchs H. E., Jemal A., Ca‐ Cancer J. Clin. 2022, 72, 7. - PubMed
-
- Cheng A.‐L., Kang Y.‐K., Chen Z., Tsao C.‐J., Qin S., Kim J. S., Luo R., Feng J., Ye S., Yang T.‐S., Xu J., Sun Y., Liang H., Liu J., Wang J., Tak W. Y., Pan H., Burock K., Zou J., Voliotis D., Guan Z., Lancet Oncol. 2009, 10, 25. - PubMed
-
- Bruix J., Qin S., Merle P., Granito A., Huang Y. H., Bodoky G., Pracht M., Yokosuka O., Rosmorduc O., Breder V., Gerolami R., Masi G., Ross P. J., Song T., Bronowicki J. P., Ollivier‐Hourmand I., Kudo M., Cheng A. L., Llovet J. M., Finn R. S., LeBerre M. A., Baumhauer A., Meinhardt G., Han G., Investigators R., Lancet 2017, 389, 56. - PubMed
-
- Bruix J., da Fonseca L. G., Reig M., Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 617. - PubMed
-
- Llovet S. R. J. M., Mazzaferro V., Hilgard P., Gane E., Blanc J. F., Oliveira A. C., Santoro A., Raoul J. L., Forner A., Schwartz M., Porta C., Zeuzem S., Bolondi L., Greten T. F., Galle P. R., Seitz J. F., Borbath I., Häussinger D., Giannaris T., Shan M., Moscovici M., Voliotis D., Bruix J., N. Engl. J. Med. 2008, 359, 378. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
