The impact of mRNA poly(A) tail length on eukaryotic translation stages
- PMID: 38874498
- PMCID: PMC11260481
- DOI: 10.1093/nar/gkae510
The impact of mRNA poly(A) tail length on eukaryotic translation stages
Abstract
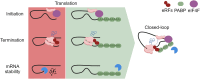
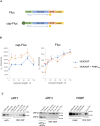
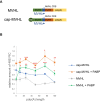
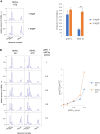
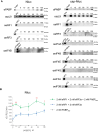
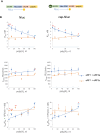
The poly(A) tail plays an important role in maintaining mRNA stability and influences translation efficiency via binding with PABP. However, the impact of poly(A) tail length on mRNA translation remains incompletely understood. This study explores the effects of poly(A) tail length on human translation. We determined the translation rates in cell lysates using mRNAs with different poly(A) tails. Cap-dependent translation was stimulated by the poly(A) tail, however, it was largely independent of poly(A) tail length, with an exception observed in the case of the 75 nt poly(A) tail. Conversely, cap-independent translation displayed a positive correlation with poly(A) tail length. Examination of translation stages uncovered the dependence of initiation and termination on the presence of the poly(A) tail, but the efficiency of initiation remained unaffected by poly(A) tail extension. Further study unveiled that increased binding of eRFs to the ribosome with the poly(A) tail extension induced more efficient hydrolysis of peptidyl-tRNA. Building upon these findings, we propose a crucial role for the 75 nt poly(A) tail in orchestrating the formation of a double closed-loop mRNA structure within human cells which couples the initiation and termination phases of translation.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Eckmann C.R., Rammelt C., Wahle E Control of poly(A) tail length. Wiley Interdiscip. Rev. RNA. 2011; 2:348–361. - PubMed
-
- Legnini I., Alles J., Karaiskos N., Ayoub S., Rajewsky N. FLAM-seq: full-length mRNA sequencing reveals principles of poly(A) tail length control. Nat. Methods. 2019; 16:879–886. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

