Exploring a novel GH13_5 α-amylase from Jeotgalibacillus malaysiensis D5T for raw starch hydrolysis
- PMID: 38874807
- PMCID: PMC11178733
- DOI: 10.1186/s13568-024-01722-3
Exploring a novel GH13_5 α-amylase from Jeotgalibacillus malaysiensis D5T for raw starch hydrolysis
Abstract
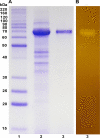
α-Amylase plays a crucial role in the industrial degradation of starch. The genus Jeotgalibacillus of the underexplored marine bacteria family Caryophanaceae has not been investigated in terms of α-amylase production. Herein, we report the comprehensive analysis of an α-amylase (AmyJM) from Jeotgalibacillus malaysiensis D5T (= DSM28777T = KCTC33550T). Protein phylogenetic analysis indicated that AmyJM belongs to glycoside hydrolase family 13 subfamily 5 (GH13_5) and exhibits low sequence identity with known α-amylases, with its closest counterpart being the GH13_5 α-amylase from Bacillus sp. KSM-K38 (51.05% identity). Purified AmyJM (molecular mass of 70 kDa) is stable at a pH range of 5.5-9.0 and optimally active at pH 7.5. The optimum temperature for AmyJM is 40 °C, where the enzyme is reasonably stable at this temperature. Similar to other α-amylases, the presence of CaCl2 enhanced both the activity and stability of AmyJM. AmyJM exhibited activity toward raw and gelatinized forms of starches and related α-glucans, generating a mixture of reducing sugars, such as glucose, maltose, maltotriose, maltotetraose, and maltopentaose. In raw starch hydrolysis, AmyJM exhibited its highest efficiency (51.10% degradation) in hydrolyzing raw wheat starch after 3-h incubation at 40 °C. Under the same conditions, AmyJM also hydrolyzed tapioca, sago, potato, rice, and corn raw starches, yielding 16.01-30.05%. These findings highlight the potential of AmyJM as a biocatalyst for the saccharification of raw starches, particularly those derived from wheat.
Keywords: Caryophanaceae; Jeotgalibacillus; Glycoside hydrolase family 13 subfamily 5; Marine bacteria; Raw starch hydrolysis; α-Amylase.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
AmyZ1: a novel α-amylase from marine bacterium Pontibacillus sp. ZY with high activity toward raw starches.Biotechnol Biofuels. 2019 Apr 23;12:95. doi: 10.1186/s13068-019-1432-9. eCollection 2019. Biotechnol Biofuels. 2019. PMID: 31044008 Free PMC article.
-
Preferential and rapid degradation of raw rice starch by an α-amylase of glycoside hydrolase subfamily GH13_37.Appl Microbiol Biotechnol. 2012 Jun;94(6):1577-84. doi: 10.1007/s00253-012-4114-0. Epub 2012 May 6. Appl Microbiol Biotechnol. 2012. PMID: 22562167
-
Isolation, expression, and characterization of raw starch degrading α-amylase from a marine lake Bacillus megaterium NL3.Heliyon. 2020 Dec 26;6(12):e05796. doi: 10.1016/j.heliyon.2020.e05796. eCollection 2020 Dec. Heliyon. 2020. PMID: 33426327 Free PMC article.
-
Identification and Characterization of Novel Malto-Oligosaccharide-Forming Amylase AmyCf from Cystobacter sp. Strain CF23.Foods. 2023 Sep 19;12(18):3487. doi: 10.3390/foods12183487. Foods. 2023. PMID: 37761198 Free PMC article.
-
α-Amylase: an enzyme specificity found in various families of glycoside hydrolases.Cell Mol Life Sci. 2014 Apr;71(7):1149-70. doi: 10.1007/s00018-013-1388-z. Epub 2013 Jun 27. Cell Mol Life Sci. 2014. PMID: 23807207 Free PMC article. Review.
Cited by
-
Characterization of a Thermostable α-Amylase from Bacillus licheniformis 104.K for Industrial Applications.Microorganisms. 2025 Jul 28;13(8):1757. doi: 10.3390/microorganisms13081757. Microorganisms. 2025. PMID: 40871262 Free PMC article.
-
Evaluation of camelina seed pods as a novel feed ingredient for ruminants: nutritional value, fermentation characteristics and nutrient digestibility.Vet Anim Sci. 2025 Jul 11;29:100477. doi: 10.1016/j.vas.2025.100477. eCollection 2025 Sep. Vet Anim Sci. 2025. PMID: 40718160 Free PMC article.
References
-
- Agirre J, Moroz O, Meier S, Brask J, Munch A, Hoff T, Andersen C, Wilson K, Davies G. The structure of the AliC GH13 α-amylase from Alicyclobacillus sp. reveals the accommodation of starch branching points in the α-amylase family. Acta Crystallogr D Struct Biol. 2019;75:1–7. doi: 10.1107/S2059798318014900. - DOI - PMC - PubMed
-
- Alikhajeh J, Khajeh K, Ranjbar B, Naderi-Manesh H, Lin Y, Liu E, Guan H, Hsieh Y, Chuankhayan P, Huang Y, Jeyaraman J, Liu M, Chen C. Structure of Bacillus amyloliquefaciens α-amylase at high resolution: implications for thermal stability. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66:121–129. doi: 10.1107/S1744309109051938. - DOI - PMC - PubMed
-
- Bertoft E. Understanding starch structure: recent progress. Agronomy. 2017;7:56. doi: 10.3390/agronomy7030056. - DOI
-
- Božić N, Lončar N, Slavić M, Vujčić Z. Raw starch degrading α-amylases: an unsolved riddle. Amylase. 2017;1:12–25. doi: 10.1515/amylase-2017-0002. - DOI
LinkOut - more resources
Full Text Sources
Research Materials

