microRNA-guided immunity against respiratory virus infection in human and mouse lung cells
- PMID: 38875000
- PMCID: PMC11212637
- DOI: 10.1242/bio.060172
microRNA-guided immunity against respiratory virus infection in human and mouse lung cells
Abstract
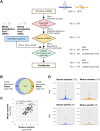
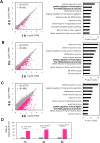
Viral infectivity depends on multiple factors. Recent studies showed that the interaction between viral RNAs and endogenous microRNAs (miRNAs) regulates viral infectivity; viral RNAs function as a sponge of endogenous miRNAs and result in upregulation of its original target genes, while endogenous miRNAs target viral RNAs directly and result in repression of viral gene expression. In this study, we analyzed the possible interaction between parainfluenza virus RNA and endogenous miRNAs in human and mouse lungs. We showed that the parainfluenza virus can form base pairs with human miRNAs abundantly than mouse miRNAs. Furthermore, we analyzed that the sponge effect of endogenous miRNAs on viral RNAs may induce the upregulation of transcription regulatory factors. Then, we performed RNA-sequence analysis and observed the upregulation of transcription regulatory factors in the early stages of parainfluenza virus infection. Our studies showed how the differential expression of endogenous miRNAs in lungs could contribute to respiratory virus infection and species- or tissue-specific mechanisms and common mechanisms could be conserved in humans and mice and regulated by miRNAs during viral infection.
Keywords: Lungs; MicroRNA; RNA silencing; RNA–RNA interaction; Respiratory virus.
© 2024. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures




References
-
- Alberts, B., Heald, R., Johnson, A. and Morgan, D. (2022). Molecular Biology of the Cell. New York City, NY: W.W. Norton & Co Inc.
-
- Bartoszewski, R., Dabrowski, M., Jakiela, B., Matalon, S., Harrod, K. S., Sanak, M. and Collawn, J. F. (2020). SARS-CoV-2 may regulate cellular responses through depletion of specific host miRNAs. Am. J. Physiol. Lung Cell. Mol. Physiol. 319, L444-L455. 10.1152/ajplung.00252.2020 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

