Antiviral activity of intracellular nanobodies targeting the influenza virus RNA-polymerase core
- PMID: 38875296
- PMCID: PMC11210859
- DOI: 10.1371/journal.ppat.1011642
Antiviral activity of intracellular nanobodies targeting the influenza virus RNA-polymerase core
Abstract
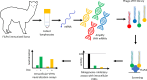
Influenza viruses transcribe and replicate their genome in the nucleus of the infected cells, two functions that are supported by the viral RNA-dependent RNA-polymerase (FluPol). FluPol displays structural flexibility related to distinct functional states, from an inactive form to conformations competent for replication and transcription. FluPol machinery is constituted by a structurally-invariant core comprising the PB1 subunit stabilized with PA and PB2 domains, whereas the PA endonuclease and PB2 C-domains can pack in different configurations around the core. To get insights into the functioning of FluPol, we selected single-domain nanobodies (VHHs) specific of the influenza A FluPol core. When expressed intracellularly, some of them exhibited inhibitory activity on type A FluPol, but not on the type B one. The most potent VHH (VHH16) binds PA and the PA-PB1 dimer with an affinity below the nanomolar range. Ectopic intracellular expression of VHH16 in virus permissive cells blocks multiplication of different influenza A subtypes, even when induced at late times post-infection. VHH16 was found to interfere with the transport of the PA-PB1 dimer to the nucleus, without affecting its handling by the importin β RanBP5 and subsequent steps in FluPol assembly. Using FluPol mutants selected after passaging in VHH16-expressing cells, we identified the VHH16 binding site at the interface formed by PA residues with the N-terminus of PB1, overlapping or close to binding sites of two host proteins, ANP32A and RNA-polymerase II RPB1 subunit which are critical for virus replication and transcription, respectively. These data suggest that the VHH16 neutralization is likely due to several activities, altering the import of the PA-PB1 dimer into the nucleus as well as inhibiting specifically virus transcription and replication. Thus, the VHH16 binding site represents a new Achilles' heel for FluPol and as such, a potential target for antiviral development.
Copyright: © 2024 Bessonne et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
-
- EFSA (European Food Safety Authority), ECDC (European Centre for Disease Prevention and Control), EURL (European Reference Laboratory for Avian Influenza), Adlhoch C, Fusaro A, Gonzales JL, Kuiken T, Marangon S, Niqueux É, Staubach C, Terregino C, Aznar I,Chuzhakina K, Muñoz Guajardo I and Baldinelli F, 2022. Scientific report: Avian influenza overview June–September2022.EFSA Journal 2022;20(10):7597,58pp. 10.2903/j.efsa.2022.7597 - DOI
-
- Shaw M, Palese P. 2013. Orthomyxoviridae, p 1151–1185. In Knipe DM, Howley P (ed), Fields virology, 6th ed, vol 1. Lippincott Williams & Wilkins, Philadelphia, PA.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

