Using random forests to uncover the predictive power of distance-varying cell interactions in tumor microenvironments
- PMID: 38875302
- PMCID: PMC11210873
- DOI: 10.1371/journal.pcbi.1011361
Using random forests to uncover the predictive power of distance-varying cell interactions in tumor microenvironments
Abstract
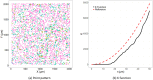
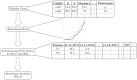
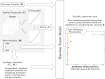
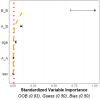
Tumor microenvironments (TMEs) contain vast amounts of information on patient's cancer through their cellular composition and the spatial distribution of tumor cells and immune cell populations. Exploring variations in TMEs between patient groups, as well as determining the extent to which this information can predict outcomes such as patient survival or treatment success with emerging immunotherapies, is of great interest. Moreover, in the face of a large number of cell interactions to consider, we often wish to identify specific interactions that are useful in making such predictions. We present an approach to achieve these goals based on summarizing spatial relationships in the TME using spatial K functions, and then applying functional data analysis and random forest models to both predict outcomes of interest and identify important spatial relationships. This approach is shown to be effective in simulation experiments at both identifying important spatial interactions while also controlling the false discovery rate. We further used the proposed approach to interrogate two real data sets of Multiplexed Ion Beam Images of TMEs in triple negative breast cancer and lung cancer patients. The methods proposed are publicly available in a companion R package funkycells.
Copyright: © 2024 VanderDoes et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
MeSH terms
LinkOut - more resources
Full Text Sources

