Interplay between Pitx2 and Pax7 temporally governs specification of extraocular muscle stem cells
- PMID: 38875306
- PMCID: PMC11178213
- DOI: 10.1371/journal.pgen.1010935
Interplay between Pitx2 and Pax7 temporally governs specification of extraocular muscle stem cells
Abstract
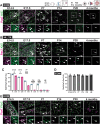
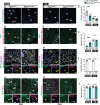
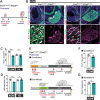
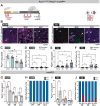
Gene regulatory networks that act upstream of skeletal muscle fate determinants are distinct in different anatomical locations. Despite recent efforts, a clear understanding of the cascade of events underlying the emergence and maintenance of the stem cell pool in specific muscle groups remains unresolved and debated. Here, we invalidated Pitx2 with multiple Cre-driver mice prenatally, postnatally, and during lineage progression. We showed that this gene becomes progressively dispensable for specification and maintenance of the muscle stem (MuSC) cell pool in extraocular muscles (EOMs) despite being, together with Myf5, a major upstream regulator during early development. Moreover, constitutive inactivation of Pax7 postnatally led to a greater loss of MuSCs in the EOMs compared to the limb. Thus, we propose a relay between Pitx2, Myf5 and Pax7 for EOM stem cell maintenance. We demonstrate also that MuSCs in the EOMs adopt a quiescent state earlier that those in limb muscles and do not spontaneously proliferate in the adult, yet EOMs have a significantly higher content of Pax7+ MuSCs per area pre- and post-natally. Finally, while limb MuSCs proliferate in the mdx mouse model for Duchenne muscular dystrophy, significantly less MuSCs were present in the EOMs of the mdx mouse model compared to controls, and they were not proliferative. Overall, our study provides a comprehensive in vivo characterisation of MuSC heterogeneity along the body axis and brings further insights into the unusual sparing of EOMs during muscular dystrophy.
Copyright: © 2024 Kuriki et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Dystrophic changes in extraocular muscles after gamma irradiation in mdx:utrophin(+/-) mice.PLoS One. 2014 Jan 21;9(1):e86424. doi: 10.1371/journal.pone.0086424. eCollection 2014. PLoS One. 2014. PMID: 24466085 Free PMC article.
-
The role of Pitx2 in maintaining the phenotype of myogenic precursor cells in the extraocular muscles.PLoS One. 2013;8(3):e58405. doi: 10.1371/journal.pone.0058405. Epub 2013 Mar 7. PLoS One. 2013. PMID: 23505501 Free PMC article.
-
Extraocular muscle satellite cells are high performance myo-engines retaining efficient regenerative capacity in dystrophin deficiency.Dev Biol. 2015 Jan 1;397(1):31-44. doi: 10.1016/j.ydbio.2014.08.035. Epub 2014 Sep 16. Dev Biol. 2015. PMID: 25236433 Free PMC article.
-
The molecular regulation of muscle stem cell function.Cold Spring Harb Symp Quant Biol. 2008;73:323-31. doi: 10.1101/sqb.2008.73.064. Epub 2009 Mar 27. Cold Spring Harb Symp Quant Biol. 2008. PMID: 19329572 Review.
-
Stem and progenitor cells in skeletal muscle development, maintenance, and therapy.Mol Ther. 2007 May;15(5):867-77. doi: 10.1038/mt.sj.6300145. Epub 2007 Mar 27. Mol Ther. 2007. PMID: 17387336 Review.
Cited by
-
Targeting Msx2 as a brake in the fusion fate of osteoclasts and an anabolic therapy in pre-clinical models of osteoporosis.Nat Commun. 2025 Aug 6;16(1):7228. doi: 10.1038/s41467-025-61938-0. Nat Commun. 2025. PMID: 40764291 Free PMC article.
References
-
- Suzuki DG, Fukumoto Y, Yoshimura M, Yamazaki Y, Kosaka J, Kuratani S, et al.. Comparative morphology and development of extra-ocular muscles in the lamprey and gnathostomes reveal the ancestral state and developmental patterns of the vertebrate head. Zoological Lett. 2016;2:10. Epub 20160414. doi: 10.1186/s40851-016-0046-3 ; PubMed Central PMCID: PMC4831119. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

