Modeling the transmission mitigation impact of testing for infectious diseases
- PMID: 38875334
- PMCID: PMC11177932
- DOI: 10.1126/sciadv.adk5108
Modeling the transmission mitigation impact of testing for infectious diseases
Abstract
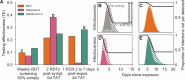
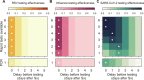
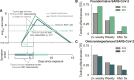
A fundamental question of any program focused on the testing and timely diagnosis of a communicable disease is its effectiveness in reducing transmission. Here, we introduce testing effectiveness (TE)-the fraction by which testing and post-diagnosis isolation reduce transmission at the population scale-and a model that incorporates test specifications and usage, within-host pathogen dynamics, and human behaviors to estimate TE. Using TE to guide recommendations, we show that today's rapid diagnostics should be used immediately upon symptom onset to control influenza A and respiratory syncytial virus but delayed by up to two days to control omicron-era severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Furthermore, while rapid tests are superior to reverse transcription quantitative polymerase chain reaction (RT-qPCR) to control founder-strain SARS-CoV-2, omicron-era changes in viral kinetics and rapid test sensitivity cause a reversal, with higher TE for RT-qPCR despite longer turnaround times. Last, we illustrate the model's flexibility by quantifying trade-offs in the use of post-diagnosis testing to shorten isolation times.
Figures


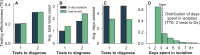
Update of
-
Modeling the Transmission Mitigation Impact of Testing for Infectious Diseases.medRxiv [Preprint]. 2024 Mar 5:2023.09.22.23295983. doi: 10.1101/2023.09.22.23295983. medRxiv. 2024. Update in: Sci Adv. 2024 Jun 14;10(24):eadk5108. doi: 10.1126/sciadv.adk5108. PMID: 37808825 Free PMC article. Updated. Preprint.
References
-
- Ranoa D. R. E., Holland R. L., Alnaji F. G., Green K. J., Wang L., Fredrickson R. L., Wang T., Wong G. N., Uelmen J., Maslov S., Weiner Z. J., Tkachenko A. V., Zhang H., Liu Z., Ibrahim A., Patel S. J., Paul J. M., Vance N. P., Gulick J. G., Satheesan S. P., Galvan I. J., Miller A., Grohens J., Nelson T. J., Stevens M. P., Hennessy P. M., Parker R. C. Jr., Santos E., Brackett C., Steinman J. D., Fenner M. R. Jr., Dohrer K., De Lorenzo M., Wilhelm-Barr L., Brauer B. R., Best-Popescu C., Durack G., Wetter N., Kranz D. M., Breitbarth J., Simpson C., Pryde J. A., Kaler R. N., Harris C., Vance A. C., Silotto J. L., Johnson M., Valera E. A., Anton P. K., Mwilambwe L., Bryan S. P., Stone D. S., Young D. B., Ward W. E., Lantz J., Vozenilek J. A., Bashir R., Moore J. S., Garg M., Cooper J. C., Snyder G., Lore M. H., Yocum D. L., Cohen N. J., Novakofski J. E., Loots M. J., Ballard R. L., Band M., Banks K. M., Barnes J. D., Bentea I., Black J., Busch J., Conte A., Conte M., Curry M., Eardley J., Edwards A., Eggett T., Fleurimont J., Foster D., Fouke B. W., Gallagher N., Gastala N., Genung S. A., Glueck D., Gray B., Greta A., Healy R. M., Hetrick A., Holterman A. A., Ismail N., Jasenof I., Kelly P., Kielbasa A., Kiesel T., Kindle L. M., Lipking R. L., Manabe Y. C., Mayes J., Guffin R. M., Henry K. G. M., Mirza A., Moseley J., Mostafa H. H., Mumford M., Munoz K., Murray A. D., Nolan M., Parikh N. A., Pekosz A., Pflugmacher J., Phillips J. M., Pitts C., Potter M. C., Quisenberry J., Rear J., Robinson M. L., Rosillo E., Rye L. N., Sherwood M. E., Simon A., Singson J. M., Skadden C., Skelton T. H., Smith C., Stech M., Thomas R., Tomaszewski M. A., Tyburski E. A., Vanwingerden S., Vlach E., Watkins R. S., Watson K., White K. C., Killeen T. L., Jones R. J., Cangellaris A. C., Martinis S. A., Vaid A., Brooke C. B., Walsh J. T., Elbanna A., Sullivan W. C., Smith R. L., Goldenfeld N., Fan T. M., Hergenrother P. J., Burke M. D., Mitigation of SARS-CoV-2 transmission at a large public university. Nat. Commun. 13, 3207 (2022). - PMC - PubMed
-
- Merckx J., Wali R., Schiller I., Caya C., Gore G. C., Chartrand C., Dendukuri N., Papenburg J., Diagnostic accuracy of novel and traditional rapid tests for influenza infection compared with reverse transcriptase polymerase chain reaction: A systematic review and meta-analysis. Ann. Intern. Med. 167, 394–409 (2017). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

