Single-cell and spatially resolved interactomics of tooth-associated keratinocytes in periodontitis
- PMID: 38876998
- PMCID: PMC11178863
- DOI: 10.1038/s41467-024-49037-y
Single-cell and spatially resolved interactomics of tooth-associated keratinocytes in periodontitis
Abstract
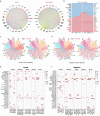
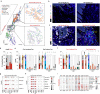
Periodontitis affects billions of people worldwide. To address relationships of periodontal niche cell types and microbes in periodontitis, we generated an integrated single-cell RNA sequencing (scRNAseq) atlas of human periodontium (34-sample, 105918-cell), including sulcular and junctional keratinocytes (SK/JKs). SK/JKs displayed altered differentiation states and were enriched for effector cytokines in periodontitis. Single-cell metagenomics revealed 37 bacterial species with cell-specific tropism. Fluorescence in situ hybridization detected intracellular 16 S and mRNA signals of multiple species and correlated with SK/JK proinflammatory phenotypes in situ. Cell-cell communication analysis predicted keratinocyte-specific innate and adaptive immune interactions. Highly multiplexed immunofluorescence (33-antibody) revealed peri-epithelial immune foci, with innate cells often spatially constrained around JKs. Spatial phenotyping revealed immunosuppressed JK-microniches and SK-localized tertiary lymphoid structures in periodontitis. Here, we demonstrate impacts on and predicted interactomics of SK and JK cells in health and periodontitis, which requires further investigation to support precision periodontal interventions in states of chronic inflammation.
© 2024. The Author(s).
Conflict of interest statement
The authors had access to the study data and reviewed and approved the final manuscript. Although the authors view each of these as noncompeting financial interests, K.M.B., Q.T.E., B.F.M., D.P., T.W., A.H., I.S., J.L., and S.A.T. are all active members of the Human Cell Atlas; furthermore, K.M.B. is a scientific advisor at Arcato Laboratories; I.S. a consultant for L’Oréal Research and Innovation; and S.A.T. has consulted for Roche and Genentech and is a scientific advisor for Biogen, GlaxoSmithKline, and Foresite Labs. All other authors declare no competing interests.
Figures






References
-
- Jepsen S, et al. Periodontal manifestations of systemic diseases and developmental and acquired conditions: consensus report of workgroup 3 of the 2017 world workshop on the classification of periodontal and peri‐implant diseases and conditions. J. Clin. Periodontol. 2018;45:S219–S229. doi: 10.1111/jcpe.12951. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

