Cardiac myosin binding protein-C phosphorylation as a function of multiple protein kinase and phosphatase activities
- PMID: 38877002
- PMCID: PMC11178824
- DOI: 10.1038/s41467-024-49408-5
Cardiac myosin binding protein-C phosphorylation as a function of multiple protein kinase and phosphatase activities
Abstract
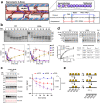
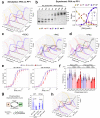
Phosphorylation of cardiac myosin binding protein-C (cMyBP-C) is a determinant of cardiac myofilament function. Although cMyBP-C phosphorylation by various protein kinases has been extensively studied, the influence of protein phosphatases on cMyBP-C's multiple phosphorylation sites has remained largely obscure. Here we provide a detailed biochemical characterization of cMyBP-C dephosphorylation by protein phosphatases 1 and 2 A (PP1 and PP2A), and develop an integrated kinetic model for cMyBP-C phosphorylation using data for both PP1, PP2A and various protein kinases known to phosphorylate cMyBP-C. We find strong site-specificity and a hierarchical mechanism for both phosphatases, proceeding in the opposite direction of sequential phosphorylation by potein kinase A. The model is consistent with published data from human patients and predicts complex non-linear cMyBP-C phosphorylation patterns that are validated experimentally. Our results suggest non-redundant roles for PP1 and PP2A under both physiological and heart failure conditions, and emphasize the importance of phosphatases for cMyBP-C regulation.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Cardiac myosin-binding protein C N-terminal interactions with myosin and actin filaments: Opposite effects of phosphorylation and M-domain mutations.J Mol Cell Cardiol. 2024 Jan;186:125-137. doi: 10.1016/j.yjmcc.2023.11.010. Epub 2023 Nov 24. J Mol Cell Cardiol. 2024. PMID: 38008210 Free PMC article.
-
The HCM-linked W792R mutation in cardiac myosin-binding protein C reduces C6 FnIII domain stability.Am J Physiol Heart Circ Physiol. 2018 Jun 1;314(6):H1179-H1191. doi: 10.1152/ajpheart.00686.2017. Epub 2018 Feb 16. Am J Physiol Heart Circ Physiol. 2018. PMID: 29451820 Free PMC article.
-
Elucidation of roles of serine/threonine phosphatases PP1 and PP2A in mediating CCK-stimulated growth and enzyme secretion in pancreatic acinar cells.Am J Physiol Gastrointest Liver Physiol. 2025 Jul 1;329(1):G102-G121. doi: 10.1152/ajpgi.00308.2024. Epub 2025 May 15. Am J Physiol Gastrointest Liver Physiol. 2025. PMID: 40375576 Free PMC article.
-
Antidepressants for pain management in adults with chronic pain: a network meta-analysis.Health Technol Assess. 2024 Oct;28(62):1-155. doi: 10.3310/MKRT2948. Health Technol Assess. 2024. PMID: 39367772 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
Cited by
-
GelBox: open-source software to improve rigor and reproducibility when analyzing gels and immunoblots.Am J Physiol Heart Circ Physiol. 2024 Sep 1;327(3):H715-H721. doi: 10.1152/ajpheart.00144.2024. Epub 2024 Aug 2. Am J Physiol Heart Circ Physiol. 2024. PMID: 39092999 Free PMC article.
-
Smart responsive biomaterials for spatiotemporal modulation of functional tissue repair.Mater Today Bio. 2025 Jul 9;33:102063. doi: 10.1016/j.mtbio.2025.102063. eCollection 2025 Aug. Mater Today Bio. 2025. PMID: 40697324 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

