The effector-triggered immunity landscape of tomato against Pseudomonas syringae
- PMID: 38877009
- PMCID: PMC11178782
- DOI: 10.1038/s41467-024-49425-4
The effector-triggered immunity landscape of tomato against Pseudomonas syringae
Abstract
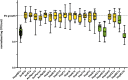
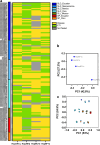
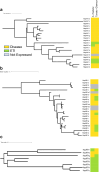
Tomato (Solanum lycopersicum) is one of the world's most important food crops, and as such, its production needs to be protected from infectious diseases that can significantly reduce yield and quality. Here, we survey the effector-triggered immunity (ETI) landscape of tomato against the bacterial pathogen Pseudomonas syringae. We perform comprehensive ETI screens in five cultivated tomato varieties and two wild relatives, as well as an immunodiversity screen on a collection of 149 tomato varieties that includes both wild and cultivated varieties. The screens reveal a tomato ETI landscape that is more limited than what was previously found in the model plant Arabidopsis thaliana. We also demonstrate that ETI eliciting effectors can protect tomato against P. syringae infection when the effector is delivered by a non-virulent strain either prior to or simultaneously with a virulent strain. Overall, our findings provide a snapshot of the ETI landscape of tomatoes and demonstrate that ETI can be used as a biocontrol treatment to protect crop plants.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Effector-triggered immunity mediated by the Pto kinase.Trends Plant Sci. 2011 Mar;16(3):132-40. doi: 10.1016/j.tplants.2010.11.001. Epub 2010 Nov 26. Trends Plant Sci. 2011. PMID: 21112235 Review.
-
HopAS1 recognition significantly contributes to Arabidopsis nonhost resistance to Pseudomonas syringae pathogens.New Phytol. 2012 Jan;193(1):58-66. doi: 10.1111/j.1469-8137.2011.03950.x. Epub 2011 Nov 4. New Phytol. 2012. PMID: 22053875
-
The pan-genome effector-triggered immunity landscape of a host-pathogen interaction.Science. 2020 Feb 14;367(6479):763-768. doi: 10.1126/science.aax4079. Science. 2020. PMID: 32054757
-
The Pseudomonas syringae type III effector HopG1 targets mitochondria, alters plant development and suppresses plant innate immunity.Cell Microbiol. 2010 Mar;12(3):318-30. doi: 10.1111/j.1462-5822.2009.01396.x. Epub 2009 Oct 27. Cell Microbiol. 2010. PMID: 19863557 Free PMC article.
-
Proteomics of effector-triggered immunity (ETI) in plants.Virulence. 2014;5(7):752-60. doi: 10.4161/viru.36329. Virulence. 2014. PMID: 25513776 Free PMC article. Review.
Cited by
-
Plant Defense Proteins: Recent Discoveries and Applications.Plants (Basel). 2025 Jul 6;14(13):2069. doi: 10.3390/plants14132069. Plants (Basel). 2025. PMID: 40648078 Free PMC article. Review.
-
Identification of Chlortetracycline Hydrochloride as a Antibacterial Compound Against Pseudomonas syringae pv. tomato DC3000 Through Natural Products Library Screening.Curr Microbiol. 2025 Apr 16;82(6):245. doi: 10.1007/s00284-025-04221-1. Curr Microbiol. 2025. PMID: 40240549
-
A SlMYB78-regulated bifunctional gene cluster for phenolamide and salicylic acid biosynthesis during tomato domestication, reducing disease resistance.J Integr Plant Biol. 2025 Jul;67(7):1947-1964. doi: 10.1111/jipb.13899. Epub 2025 Mar 28. J Integr Plant Biol. 2025. PMID: 40152226 Free PMC article.
-
Utilizing effector-triggered immunity (ETI) as a robust priming agent to protect plants from pathogens.Stress Biol. 2024 Dec 9;4(1):51. doi: 10.1007/s44154-024-00204-7. Stress Biol. 2024. PMID: 39648222 Free PMC article. No abstract available.
References
-
- Peralta IE, Spooner DM. Classification of wild tomatoes: a review. Kurtziana. 2000;28:45–54.
-
- Panno S, et al. A review of the most common and economically important diseases that undermine the cultivation of tomato crop in the Mediterranean basin. Agronomy. 2021;11:2188. doi: 10.3390/agronomy11112188. - DOI
-
- LeBoeuf, J. Bacterial diseases of tomato: Bacterial spot, bacterial speck, and bacterial canker. Ontario Ministry of Agriculture, Food & Rural Affairs. https://www.ontario.ca/page/bacterial-diseases-tomato-bacterial-spot-bac... (2005).
-
- Lamichhane JR, Messéan A, Morris CE. Insights into epidemiology and control of diseases of annual plants caused by the Pseudomonas syringae species complex. J. Gen. Plant Pathol. 2015;81:331–350. doi: 10.1007/s10327-015-0605-z. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

