Ultrasensitive plasma-based monitoring of tumor burden using machine-learning-guided signal enrichment
- PMID: 38877116
- PMCID: PMC7616143
- DOI: 10.1038/s41591-024-03040-4
Ultrasensitive plasma-based monitoring of tumor burden using machine-learning-guided signal enrichment
Abstract
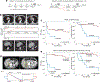
In solid tumor oncology, circulating tumor DNA (ctDNA) is poised to transform care through accurate assessment of minimal residual disease (MRD) and therapeutic response monitoring. To overcome the sparsity of ctDNA fragments in low tumor fraction (TF) settings and increase MRD sensitivity, we previously leveraged genome-wide mutational integration through plasma whole-genome sequencing (WGS). Here we now introduce MRD-EDGE, a machine-learning-guided WGS ctDNA single-nucleotide variant (SNV) and copy-number variant (CNV) detection platform designed to increase signal enrichment. MRD-EDGESNV uses deep learning and a ctDNA-specific feature space to increase SNV signal-to-noise enrichment in WGS by ~300× compared to previous WGS error suppression. MRD-EDGECNV also reduces the degree of aneuploidy needed for ultrasensitive CNV detection through WGS from 1 Gb to 200 Mb, vastly expanding its applicability within solid tumors. We harness the improved performance to identify MRD following surgery in multiple cancer types, track changes in TF in response to neoadjuvant immunotherapy in lung cancer and demonstrate ctDNA shedding in precancerous colorectal adenomas. Finally, the radical signal-to-noise enrichment in MRD-EDGESNV enables plasma-only (non-tumor-informed) disease monitoring in advanced melanoma and lung cancer, yielding clinically informative TF monitoring for patients on immune-checkpoint inhibition.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
DAL, AJW, CCK, and JB are listed as inventors on a pending patent application (WO2023018791A1), filed by Cornell University, that is directed to methods of detecting SNVs for the purposes of MRD detection and other plasma-based cancer monitoring. DAL, AJW, and MS are listed as inventors on a pending patent application (WO2023133093A1), filed by Cornell University, that is directed to methods of detecting CNVs for the purposes of MRD detection and other plasma-based cancer monitoring. APC is listed as an inventor on submitted patents pertaining to cell-free DNA (US patent applications 63/237,367, 63/056,249, 63/015,095, 16/500,929) and receives consulting fees from Eurofins Viracor. AS receives research funding from AstraZeneca, has served on Advisory Boards for AstraZeneca, Blueprint Medicines, and Jazz Pharmaceuticals, and has been a consultant for Genentech. MAP has received consulting fees from BMS, Chugai, Cancer Expert Now, Intellisphere, Merck, MJH associates, Nektar, Pfizer, Uptodate, WebMD, Erasca, and received institutional support from RGenix, Infinity, BMS, Merck, Genentech and Novartis. CLA reports collaborations with C2i Genomics and Natera. CS has received honoraria for advisory board participation from Pfizer, Novartis, Knight, Bayer, Merck, Roche, Lilly within the past two years. None of the honoraria have been in excess of $5000.00CAD and total for honoraria received is less than $25,000.00CAD. MKC has received consulting fees from BMS, Merck, InCyte, Moderna, ImmunoCore, and AstraZeneca and receives institutional support from BMS. ST is funded by Cancer Research UK (grant reference number A29911); the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC10988), the UK Medical Research Council (FC10988), and the Wellcome Trust (FC10988); the National Institute for Health Research (NIHR) Biomedical Research Centre at the Royal Marsden Hospital and Institute of Cancer Research (grant reference number A109), the Royal Marsden Cancer Charity, The Rosetrees Trust (grant reference number A2204), Ventana Medical Systems Inc (grant reference numbers 10467 and 10530), the National Institute of Health (U01 CA247439) and Melanoma Research Alliance (Award Ref no 686061). ST has received speaking fees from Roche, Astra Zeneca, Novartis and Ipsen. ST has the following patents filed: Indel mutations as a therapeutic target and predictive biomarker PCTGB2018/051892 and PCTGB2018/051893. GMB has sponsored research agreements through her institution with: Olink Proteomics, Teiko Bio, InterVenn Biosciences, Palleon Pharmaceuticals. GMB served on advisory boards for: Iovance, Merck, Nektar Therapeutics, Novartis, and Ankyra Therapeutics. GMB consults for: Merck, InterVenn Biosciences, Iovance, and Ankyra Therapeutics. GMB equity in Ankyra Therapeutics. MB reports consulting for AstraZeneca, Eli Lilly, Paige.AI, research support from Boundless Bio, and intellectual property rights for SOPHiA Genetics. JDW is a consultant for: Apricity; Ascentage Pharma; AstraZeneca; BeiGene; Bicara Therapeutics; Bristol Myers Squibb; Daiichi Sankyo; Dragonfly; Imvaq; Larkspur; Psioxus, Recepta; Takeda; Tizona; Trishula Therapeutics; Sellas. JDW received Grant/Research Support from: Bristol Myers Squibb; Enterome. JDW has Equity in: Apricity, Arsenal IO/CellCarta; Ascentage; Imvaq; Linneaus, Larkspur; Georgiamune; Maverick; Tizona Therapeutics; Xenimmune. DAL received research support from Illumina, Inc. DAL participated in advisory boards Pangea, Mission Bio, Alethiomics. DAL is a scientific co-founder of C2i Genomics. The remaining authors declare no competing interests.
Figures



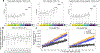




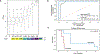





References
-
- Bratman SV et al. Personalized circulating tumor DNA analysis as a predictive biomarker in solid tumor patients treated with pembrolizumab. Nature Cancer 1, 873–881 (2020). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

